ABC-transporter CFTR folds with high fidelity through a modular, stepwise pathway
- PMID: 36609925
- PMCID: PMC9825563
- DOI: 10.1007/s00018-022-04671-x
ABC-transporter CFTR folds with high fidelity through a modular, stepwise pathway
Abstract
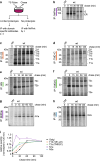
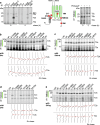
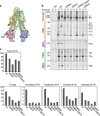
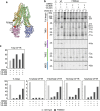
The question how proteins fold is especially pointed for large multi-domain, multi-spanning membrane proteins with complex topologies. We have uncovered the sequence of events that encompass proper folding of the ABC transporter CFTR in live cells by combining kinetic radiolabeling with protease-susceptibility assays. We found that CFTR folds in two clearly distinct stages. The first, co-translational, stage involves folding of the 2 transmembrane domains TMD1 and TMD2, plus one nucleotide-binding domain, NBD1. The second stage is a simultaneous, post-translational increase in protease resistance for both TMDs and NBD2, caused by assembly of these domains onto NBD1. Our assays probe every 2-3 residues (on average) in CFTR. This in-depth analysis at amino-acid level allows detailed analysis of domain folding and importantly also the next level: assembly of the domains into native, folded CFTR. Defects and changes brought about by medicines, chaperones, or mutations also are amenable to analysis. We here show that the well-known disease-causing mutation F508del, which established cystic fibrosis as protein-folding disease, caused co-translational misfolding of NBD1 but not TMD1 nor TMD2 in stage 1, leading to absence of stage-2 folding. Corrector drugs rescued stage 2 without rescuing NBD1. Likewise, the DxD motif in NBD1 that was identified to be required for export of CFTR from the ER we found to be required already upstream of export as CFTR mutated in this motif phenocopies F508del CFTR. The highly modular and stepwise folding process of such a large, complex protein explains the relatively high fidelity and correctability of its folding.
Keywords: ABC-transporter; COPII; Cystic fibrosis; Domain assembly; Protein folding; Secretory pathway.
© 2023. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

