Human Placental Endothelial Cell and Trophoblast Heterogeneity and Differentiation Revealed by Single-Cell RNA Sequencing
- PMID: 36611882
- PMCID: PMC9818681
- DOI: 10.3390/cells12010087
Human Placental Endothelial Cell and Trophoblast Heterogeneity and Differentiation Revealed by Single-Cell RNA Sequencing
Abstract
Background: The placenta is an important organ for fetal and maternal health during pregnancy and impacts offspring health late in life. Defects in placental vasculature and trophoblast have been identified in several pregnancy complications. Thus, the detailed molecular profile and heterogeneity of endothelial cells and trophoblasts in placentas will aid us in better understanding placental behaviors and improving pregnancy outcomes.
Methods: Single-cell RNA sequencing (scRNA-seq) was performed to profile the transcriptomics of human placental villous tissues from eleven patients with normal pregnancies in the first and second trimesters (6-16 weeks of gestation).
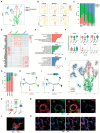
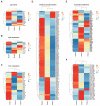
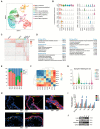
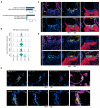
Results: The transcriptomic landscape of 52,179 single cells was obtained, and the cells were classified as trophoblasts, fibroblasts, endothelial cells, erythroid cells, Hofbauer cells, and macrophages. Our analysis further revealed the three subtypes of placental endothelial cells, with distinct metabolic signatures and transcription factor regulatory networks. We also determined the transcriptomic features of the trophoblast subpopulations and characterized two distinct populations of progenitor cells in cytotrophoblasts, which were capable of differentiating to extravillous trophoblasts and syncytiotrophoblasts, respectively.
Conclusions: Our study provided a high-resolution molecular profile of the human placenta between 6 and 16 weeks of gestation. Our data revealed the placental cell complexity and demonstrated the transcriptional networks and signaling involved in placental endothelial and trophoblast differentiation during early pregnancy, which will be a resource for future studies of the human placental development.
Keywords: endothelial cell; human placenta; pregnancy; scRNA-seq; trophoblast.
Conflict of interest statement
All authors confirm that there are no conflict of interest.
Figures


Similar articles
-
Revealing the molecular landscape of human placenta: a systematic review and meta-analysis of single-cell RNA sequencing studies.Hum Reprod Update. 2024 Jul 1;30(4):410-441. doi: 10.1093/humupd/dmae006. Hum Reprod Update. 2024. PMID: 38478759 Free PMC article.
-
Single-cell RNA-seq reveals the diversity of trophoblast subtypes and patterns of differentiation in the human placenta.Cell Res. 2018 Aug;28(8):819-832. doi: 10.1038/s41422-018-0066-y. Epub 2018 Jul 24. Cell Res. 2018. PMID: 30042384 Free PMC article.
-
Isolation and characterisation of a novel trophoblast side-population from first trimester placentae.Reproduction. 2015 Nov;150(5):449-62. doi: 10.1530/REP-14-0646. Epub 2015 Aug 6. Reproduction. 2015. PMID: 26248480
-
Sexually Dimorphic Crosstalk at the Maternal-Fetal Interface.J Clin Endocrinol Metab. 2020 Dec 1;105(12):e4831-47. doi: 10.1210/clinem/dgaa503. J Clin Endocrinol Metab. 2020. PMID: 32772088 Free PMC article.
-
Placental formation in early pregnancy: how is the centre of the placenta made?Hum Reprod Update. 2018 Nov 1;24(6):750-760. doi: 10.1093/humupd/dmy030. Hum Reprod Update. 2018. PMID: 30257012 Review.
Cited by
-
A spatiotemporal transcriptomic atlas of mouse placentation.Cell Discov. 2024 Oct 22;10(1):110. doi: 10.1038/s41421-024-00740-6. Cell Discov. 2024. PMID: 39438452 Free PMC article.
-
Lactylation of Hdac1 regulated by Ldh prevents the pluripotent-to-2C state conversion.Stem Cell Res Ther. 2024 Nov 13;15(1):415. doi: 10.1186/s13287-024-04027-1. Stem Cell Res Ther. 2024. PMID: 39533309 Free PMC article.
-
TGFβ signalling: a nexus between inflammation, placental health and preeclampsia throughout pregnancy.Hum Reprod Update. 2024 Jul 1;30(4):442-471. doi: 10.1093/humupd/dmae007. Hum Reprod Update. 2024. PMID: 38519450 Free PMC article. Review.
-
Extracellular Vesicles Alter Trophoblast Function in Pregnancies Complicated by COVID-19.J Extracell Vesicles. 2025 Apr;14(4):e70051. doi: 10.1002/jev2.70051. J Extracell Vesicles. 2025. PMID: 40205960 Free PMC article.
-
Placental single cell transcriptomics: Opportunities for endocrine disrupting chemical toxicology.Mol Cell Endocrinol. 2023 Dec 1;578:112066. doi: 10.1016/j.mce.2023.112066. Epub 2023 Sep 9. Mol Cell Endocrinol. 2023. PMID: 37690473 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

