The SMARCD Family of SWI/SNF Accessory Proteins Is Involved in the Transcriptional Regulation of Androgen Receptor-Driven Genes and Plays a Role in Various Essential Processes of Prostate Cancer
- PMID: 36611918
- PMCID: PMC9818446
- DOI: 10.3390/cells12010124
The SMARCD Family of SWI/SNF Accessory Proteins Is Involved in the Transcriptional Regulation of Androgen Receptor-Driven Genes and Plays a Role in Various Essential Processes of Prostate Cancer
Abstract
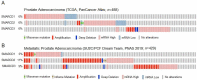
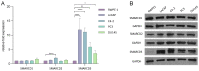
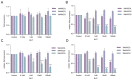
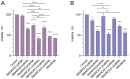
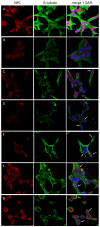
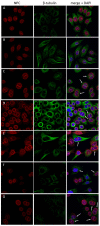
Previous studies have demonstrated an involvement of chromatin-remodelling SWI/SNF complexes in the development of prostate cancer, suggesting both tumor suppressor and oncogenic activities. SMARCD1/BAF60A, SMARCD2/BAF60B, and SMARCD3/BAF60C are mutually exclusive accessory subunits that confer functional specificity and are components of all known SWI/SNF subtypes. To assess the role of SWI/SNF in prostate tumorigenesis, we studied the functions and functional relations of the SMARCD family members. Performing RNA-seq in LnCAP cells grown in the presence or absence of dihydrotestosterone, we found that the SMARCD proteins are involved in the regulation of numerous hormone-dependent AR-driven genes. Moreover, we demonstrated that all SMARCD proteins can regulate AR-downstream targets in androgen-depleted cells, suggesting an involvement in the progression to castration-resistance. However, our approach also revealed a regulatory role for SMARCD proteins through antagonization of AR-signalling. We further demonstrated that the SMARCD proteins are involved in several important cellular processes such as the maintenance of cellular morphology and cytokinesis. Taken together, our findings suggest that the SMARCD proteins play an important, yet paradoxical, role in prostate carcinogenesis. Our approach also unmasked the complex interplay of paralogue SWI/SNF proteins that must be considered for the development of safe and efficient therapies targeting SWI/SNF.
Keywords: SMARCD1/BAF60A; SMARCD2/BAF60B; SMARCD3/BAF60C; SWI/SNF complex; chromatin-remodeling; prostate cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Low SMARCD3 expression is associated with poor prognosis in patients with prostate cancer.Prostate. 2025 Feb;85(2):181-190. doi: 10.1002/pros.24815. Epub 2024 Oct 23. Prostate. 2025. PMID: 39442954 Free PMC article.
-
Differential requirement of SWI/SNF for androgen receptor activity.J Biol Chem. 2003 Aug 15;278(33):30605-13. doi: 10.1074/jbc.M304582200. Epub 2003 May 29. J Biol Chem. 2003. PMID: 12775722
-
Targeting the BAF57 SWI/SNF subunit in prostate cancer: a novel platform to control androgen receptor activity.Cancer Res. 2008 Jun 15;68(12):4551-8. doi: 10.1158/0008-5472.CAN-07-6392. Cancer Res. 2008. PMID: 18559499 Free PMC article.
-
Advances in the role of SWI/SNF complexes in tumours.J Cell Mol Med. 2023 Apr;27(8):1023-1031. doi: 10.1111/jcmm.17709. Epub 2023 Mar 8. J Cell Mol Med. 2023. PMID: 36883311 Free PMC article. Review.
-
Opening and changing: mammalian SWI/SNF complexes in organ development and carcinogenesis.Open Biol. 2024 Oct;14(10):240039. doi: 10.1098/rsob.240039. Epub 2024 Oct 30. Open Biol. 2024. PMID: 39471843 Free PMC article. Review.
Cited by
-
miR-9-5p expression is associated with vascular invasion and prognosis in hepatocellular carcinoma, and in vitro verification.J Cancer Res Clin Oncol. 2023 Nov;149(16):14657-14671. doi: 10.1007/s00432-023-05257-1. Epub 2023 Aug 16. J Cancer Res Clin Oncol. 2023. PMID: 37584711 Free PMC article.
-
Immunotherapy, prognostic, and tumor biomarker based on pancancer analysis, SMARCD3.Aging (Albany NY). 2024 Jun 11;16(11):10074-10107. doi: 10.18632/aging.205921. Epub 2024 Jun 11. Aging (Albany NY). 2024. PMID: 38862250 Free PMC article.
-
Tumor suppressive miR-99b-5p as an epigenomic regulator mediating mTOR/AR/SMARCD1 signaling axis in aggressive prostate cancer.Front Oncol. 2023 Nov 7;13:1184186. doi: 10.3389/fonc.2023.1184186. eCollection 2023. Front Oncol. 2023. PMID: 38023145 Free PMC article.
-
Rational Design and Development of Selective BRD7 Bromodomain Inhibitors and Their Activity in Prostate Cancer.J Med Chem. 2023 Aug 24;66(16):11250-11270. doi: 10.1021/acs.jmedchem.3c00671. Epub 2023 Aug 8. J Med Chem. 2023. PMID: 37552884 Free PMC article.
-
Chromatin remodeling and cancer: the critical influence of the SWI/SNF complex.Epigenetics Chromatin. 2025 Apr 23;18(1):22. doi: 10.1186/s13072-025-00590-w. Epigenetics Chromatin. 2025. PMID: 40269969 Free PMC article. Review.
References
-
- Euskirchen G.M., Auerbach R.K., Davidov E., Gianoulis T.A., Zhong G., Rozowsky J., Bhardwaj N., Gerstein M.B., Snyder M. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 2011;7:e1002008. doi: 10.1371/journal.pgen.1002008. - DOI - PMC - PubMed
-
- Ogiwara H., Ui A., Otsuka A., Satoh H., Yokomi I., Nakajima S., Yasui A., Yokota J., Kohno T. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene. 2011;30:2135–2146. doi: 10.1038/onc.2010.592. - DOI - PubMed
-
- Hays E., Nettleton E., Carter C., Morales M., Vo L., Passo M., Vélez-Cruz R. The SWI/SNF ATPase BRG1 stimulates DNA end resection and homologous recombination by reducing nucleosome density at DNA double strand breaks and by promoting the recruitment of the CtIP nuclease. Cell Cycle. 2020;19:3096–3114. doi: 10.1080/15384101.2020.1831256. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

