Oral Microbiota as Novel Biomarkers for Colorectal Cancer Screening
- PMID: 36612188
- PMCID: PMC9818409
- DOI: 10.3390/cancers15010192
Oral Microbiota as Novel Biomarkers for Colorectal Cancer Screening
Abstract
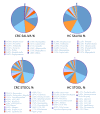
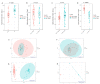
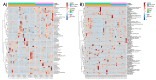
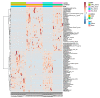
Alterations of the gut microbiome in cases of colorectal cancer (CRC) hint at the involvement of host-microbe interactions in the onset and progression of CRC and also, possibly, provide novel ways to detect and prevent CRC early. The aim of the present study was to evaluate whether the oral and fecal microbiomes of an individual can be suitable for CRC screening. Oral and fecal samples (n = 80) were gathered in Taleghani hospital, affiliated with Shahid Beheshti University of Medical Sciences, Tehran-Iran, from CRC stage 0 and I patients and healthy controls (HCs), who were screened for the first time. Microbial metagenomics assays were performed for studying microbiota profiles in all oral and fecal samples gathered. An abundance of top bacterial genera from both types of specimens (fecal and saliva samples) revealed a distinction between CRC patients and HCs. In saliva samples, the α diversity index was different between the microbiome of HCs and CRC patients, while β diversity showed a densely clustered microbiome in the HCs but a more dispersed pattern in CRC cases. The α and β diversity of fecal microbiota between HCs and CRC patients showed no statistically significant differences. Bifidobacterium was identified as a potential bacterial biomarker in CRC saliva samples, while Fusobacterium, Dialister, Catonella, Tennerella, Eubacterium-brachy-group, and Fretibacterium were ideal to distinguish HCs from CRC patients. One of the reasons for the heterogeneity of CRC may be the gastrointestinal (GI) tract microbiota, which can also cause systematic resistance to CRC. Moreover, an evaluation of saliva microbiota might offer a suitable screening test for the early detection of this malignancy, providing more accurate results than its fecal counterpart.
Keywords: biomarkers; colorectal cancer; metagenomics; non-invasive diagnosis; oral microbiota.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
LinkOut - more resources
Full Text Sources

