Iron Dyshomeostasis in COVID-19: Biomarkers Reveal a Functional Link to 5-Lipoxygenase Activation
- PMID: 36613462
- PMCID: PMC9819889
- DOI: 10.3390/ijms24010015
Iron Dyshomeostasis in COVID-19: Biomarkers Reveal a Functional Link to 5-Lipoxygenase Activation
Abstract
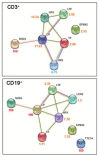
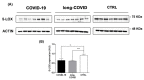
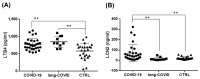
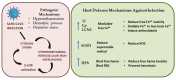
Coronavirus disease 2019 (COVID-19) is characterized by a broad spectrum of clinical symptoms. After acute infection, some subjects develop a post-COVID-19 syndrome known as long-COVID. This study aims to recognize the molecular and functional mechanisms that occur in COVID-19 and long-COVID patients and identify useful biomarkers for the management of patients with COVID-19 and long-COVID. Here, we profiled the response to COVID-19 by performing a proteomic analysis of lymphocytes isolated from patients. We identified significant changes in proteins involved in iron metabolism using different biochemical analyses, considering ceruloplasmin (Cp), transferrin (Tf), hemopexin (HPX), lipocalin 2 (LCN2), and superoxide dismutase 1 (SOD1). Moreover, our results show an activation of 5-lipoxygenase (5-LOX) in COVID-19 and in long-COVID possibly through an iron-dependent post-translational mechanism. Furthermore, this work defines leukotriene B4 (LTB4) and lipocalin 2 (LCN2) as possible markers of COVID-19 and long-COVID and suggests novel opportunities for prevention and treatment.
Keywords: 5-lipoxygenase; COVID-19; iron metabolism; leukotriene B4; lipocalin 2; long-COVID.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- World Health Organisation (WHO) COVID-19 Weekly Epidemiological Update. WHO; Geneva, Switzerland: 2022. pp. 1–26.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

