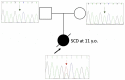
De Novo Asp219Val Mutation in Cardiac Tropomyosin Associated with Hypertrophic Cardiomyopathy
- PMID: 36613463
- PMCID: PMC9820293
- DOI: 10.3390/ijms24010018
De Novo Asp219Val Mutation in Cardiac Tropomyosin Associated with Hypertrophic Cardiomyopathy
Abstract
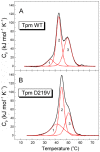
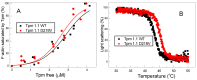
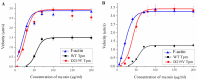
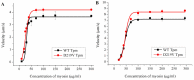
Hypertrophic cardiomyopathy (HCM), caused by mutations in thin filament proteins, manifests as moderate cardiac hypertrophy and is associated with sudden cardiac death (SCD). We identified a new de novo variant, c.656A>T (p.D219V), in the TPM1 gene encoding cardiac tropomyosin 1.1 (Tpm) in a young SCD victim with post-mortem-diagnosed HCM. We produced recombinant D219V Tpm1.1 and studied its structural and functional properties using various biochemical and biophysical methods. The D219V mutation did not affect the Tpm affinity for F-actin but increased the thermal stability of the Tpm molecule and Tpm-F-actin complex. The D219V mutation significantly increased the Ca2+ sensitivity of the sliding velocity of thin filaments over cardiac myosin in an in vitro motility assay and impaired the inhibition of the filament sliding at low Ca2+ concentration. The molecular dynamics (MD) simulation provided insight into a possible molecular mechanism of the effect of the mutation that is most likely a cause of the weakening of the Tpm interaction with actin in the "closed" state and so makes it an easier transition to the “open” state. The changes in the Ca2+ regulation of the actin-myosin interaction characteristic of genetic HCM suggest that the mutation is likely pathogenic.
Keywords: cardiomyopathic mutations; differential scanning calorimetry; hypertrophic cardiomyopathy; in vitro motility assay; molecular dynamics; tropomyosin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Coppini R., Ho C.Y., Ashley E., Day S., Ferrantini C., Girolami F., Tomberli B., Bardi S., Torricelli F., Cecchi F., et al. Clinical phenotype and outcome of hypertrophic cardiomyopathy associated with thin-filament gene mutations. J. Am. Coll. Cardiol. 2014;64:2589–2600. doi: 10.1016/j.jacc.2014.09.059. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

