Memory Macrophages
- PMID: 36613481
- PMCID: PMC9819859
- DOI: 10.3390/ijms24010038
Memory Macrophages
Abstract
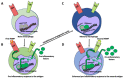
Immunological memory is a crucial part of the immune defense that allows organisms to respond against previously encountered pathogens or other harmful factors. Immunological memory is based on the establishment of epigenetic modifications of the genome. The ability to memorize encounters with pathogens and other harmful factors and mount enhanced defense upon subsequent encounters is an evolutionarily ancient mechanism operating in all animals and plants. However, the term immunological memory is usually restricted to the organisms (invertebrates and vertebrates) possessing the immune system. The mammalian immune system, with innate and adaptive branches, is the most sophisticated among vertebrates. The concept of innate memory and memory macrophages is relatively new and thus understudied. We introduce the concept of immunological memory and describe types of memory in different species and their evolutionary status. We discuss why the traditional view of innate immune cells as the first-line defenders is too restrictive and how the innate immune cells can accumulate and retain immunologic memory. We describe how the initial priming leads to chromatin remodeling and epigenetic changes, which allow memory macrophage formation. We also summarize what is currently known about the mechanisms underlying development of memory macrophages; their molecular and metabolic signature and surface markers; and how they may contribute to immune defense, diseases, and organ transplantation.
Keywords: epigenetic modifications; innate immunological memory; macrophage; trained immunity; transplantation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Janeway C.A., Jr., Travers P., Walport M., Shlomchik M.J. Immunobiology: The Immune System in Health and Disease. 5th ed. Garland Science; New York, NY, USA: 2001. [(accessed on 1 December 2022)]. Immunological Memory. Available online: https://www.ncbi.nlm.nih.gov/books/NBK27158/
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

