Gene Expression Analysis in gla-Mutant Zebrafish Reveals Enhanced Ca2+ Signaling Similar to Fabry Disease
- PMID: 36613802
- PMCID: PMC9820748
- DOI: 10.3390/ijms24010358
Gene Expression Analysis in gla-Mutant Zebrafish Reveals Enhanced Ca2+ Signaling Similar to Fabry Disease
Abstract
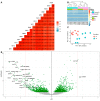
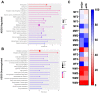
Fabry disease (FD) is an X-linked inborn metabolic disorder due to partial or complete lysosomal α-galactosidase A deficiency. FD is characterized by progressive renal insufficiency and cardio- and cerebrovascular involvement. Restricted access on Gb3-independent tissue injury experimental models has limited the understanding of FD pathophysiology and delayed the development of new therapies. Accumulating glycosphingolipids, mainly Gb3 and lysoGb3, are Fabry specific markers used in clinical follow up. However, recent studies suggest there is a need for additional markers to monitor FD clinical course or response to treatment. We used a gla-knockout zebrafish (ZF) to investigate alternative biomarkers in Gb3-free-conditions. RNA sequencing was used to identify transcriptomic signatures in kidney tissues discriminating gla-mutant (M) from wild type (WT) ZF. Gene Ontology (GO) and KEGG pathways analysis showed upregulation of immune system activation and downregulation of oxidative phosphorylation pathways in kidneys from M ZF. In addition, upregulation of the Ca2+ signaling pathway was also detectable in M ZF kidneys. Importantly, disruption of mitochondrial and lysosome-related pathways observed in M ZF was validated by immunohistochemistry. Thus, this ZF model expands the pathophysiological understanding of FD, the Gb3-independent effects of gla mutations could be used to explore new therapeutic targets for FD.
Keywords: Fabry disease; alpha-galactosidase A; calcium signaling; cardiac involvement; gla; oxidative stress; zebrafish.
Conflict of interest statement
Einar Svarstad; speaker’s fees and travel support from Amicus, Sanofi Genzyme, and Shire; advisory board honoraria from Amicus and Sanofi Genzyme. Camilla Tøndel; consultancy honoraria and/or research support from Amicus, Sanofi Genzyme, Chiesi/Protalix, Idorsia, Acelink and Freeline.
Figures


References
-
- Smid B.E., van der Tol L., Cecchi F., Elliott P.M., Hughes D.A., Linthorst G.E., Timmermans J., Weidemann F., West M.L., Biegstraaten M., et al. Uncertain diagnosis of Fabry disease: Consensus recommendation on diagnosis in adults with left ventricular hypertrophy and genetic variants of unknown significance. Int. J. Cardiol. 2014;177:400–408. doi: 10.1016/j.ijcard.2014.09.001. - DOI - PubMed
-
- Carnicer-Caceres C., Arranz-Amo J.A., Cea-Arestin C., Camprodon-Gomez M., Moreno-Martinez D., Lucas-Del-Pozo S., Molto-Abad M., Tigri-Santina A., Agraz-Pamplona I., Rodriguez-Palomares J.F., et al. Biomarkers in Fabry Disease. Implications for clinical diagnosis and follow-up. J. Clin. Med. 2021;10:1664. doi: 10.3390/jcm10081664. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

