The Efficacy of Molecular Analysis in the Diagnosis of Bone and Soft Tissue Sarcoma: A 15-Year Mono-Institutional Study
- PMID: 36614077
- PMCID: PMC9820733
- DOI: 10.3390/ijms24010632
The Efficacy of Molecular Analysis in the Diagnosis of Bone and Soft Tissue Sarcoma: A 15-Year Mono-Institutional Study
Abstract
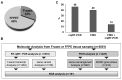
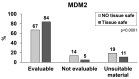
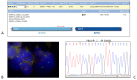
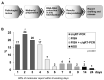
The histological diagnosis of sarcoma can be difficult as it sometimes requires the combination of morphological and immunophenotypic analyses with molecular tests. A total of 2705 tissue samples of sarcoma consecutively collected from 2006 until 2020 that had undergone molecular analysis were assessed to evaluate their diagnostic utility compared with histological assessments. A total of 3051 molecular analyses were performed, including 1484 gene fusions tested by c/qRT-PCR, 992 gene rearrangements analysed by FISH, 433 analyses of the gene status of MDM2, 126 mutational analyses and 16 NGS analysis. Of the samples analysed, 68% were from formalin-fixed, paraffin-embedded tissue and 32% were from frozen tissue. C/qRT-PCR and FISH analyses were conclusive on formalin-fixed, paraffin-embedded tissue in 74% and 76% of samples, respectively, but the combination of the two methods gave us conclusive results in 96% and 89% of frozen and formalin-fixed, paraffin-embedded tissues, respectively. We demonstrate the utility of c/qRT-PCR and FISH for sarcoma diagnosis and that each has advantages in specific contexts. We conclude that it is possible to accurately predict the sarcoma subtype using a panel of different subtype-specific FISH probes and c/qRT-PCR assays, thereby greatly facilitating the differential diagnosis of these tumours.
Keywords: FISH; RT-PCR; formalin-fixed, paraffin-embedded tissue; frozen tissue; fusion transcript; molecular diagnostics; next-generation sequencing; qRT-PCR; sarcomas.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

References
-
- Antonescu C.R., Bridge J.A., Cunha I.W. WHO Classification of Tumours Editorial Board: Soft Tissue and Bone Tumours. 5th ed. International Agency for Research on Cancer; Lyon, France: 2020. Soft tissue tumours. (WHO Classification of Tumours Series).
-
- Bovée J.V.M.G., Flanagan A.M., Lazar A.J., Nielsen G.P., Yoshida A. WHO Classification of Tumours Editorial Board: Soft Tissue and Bone Tumours. 5th ed. International Agency for Research on Cancer; Lyon, France: 2020. Bone tumours. (WHO Classification of Tumours Series).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

