Comprehensive Evaluation of Multiple Approaches Targeting ABCB1 to Resensitize Docetaxel-Resistant Prostate Cancer Cell Lines
- PMID: 36614114
- PMCID: PMC9820728
- DOI: 10.3390/ijms24010666
Comprehensive Evaluation of Multiple Approaches Targeting ABCB1 to Resensitize Docetaxel-Resistant Prostate Cancer Cell Lines
Abstract
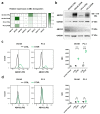
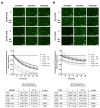
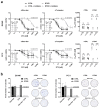
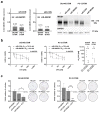
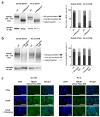
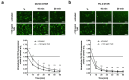
Docetaxel (DTX) is a mainstay in the treatment of metastatic prostate cancer. Failure of DTX therapy is often associated with multidrug resistance caused by overexpression of efflux membrane transporters of the ABC family such as the glycoprotein ABCB1. This study investigated multiple approaches targeting ABCB1 to resensitize DTX-resistant (DTXR) prostate cancer cell lines. In DU145 DTXR and PC-3 DTXR cells as well as age-matched parental controls, the expression of selected ABC transporters was analyzed by quantitative PCR, Western blot, flow cytometry and immunofluorescence. ABCB1 effluxing activity was studied using the fluorescent ABCB1 substrate rhodamine 123. The influence of ABCB1 inhibitors (elacridar, tariquidar), ABCB1-specific siRNA and inhibition of post-translational glycosylation on DTX tolerance was assessed by cell viability and colony formation assays. In DTXR cells, only ABCB1 was highly upregulated, which was accompanied by a strong effluxing activity and additional post-translational glycosylation of ABCB1. Pharmacological inhibition and siRNA-mediated knockdown of ABCB1 completely resensitized DTXR cells to DTX. Inhibition of glycosylation with tunicamycin affected DTX resistance partially in DU145 DTXR cells, which was accompanied by a slight intracellular accumulation and decreased effluxing activity of ABCB1. In conclusion, DTX resistance can be reversed by various strategies with small molecule inhibitors representing the most promising and feasible approach.
Keywords: ABCB1; P-glycoprotein; chemoresistance; docetaxel; elacridar; glycosylation; prostate cancer; siRNA; tariquidar; tunicamycin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Overcoming ABCB1 mediated multidrug resistance in castration resistant prostate cancer.Cell Death Dis. 2024 Aug 1;15(8):558. doi: 10.1038/s41419-024-06949-3. Cell Death Dis. 2024. PMID: 39090086 Free PMC article.
-
Protein Disulfide Isomerase 4 Drives Docetaxel Resistance in Prostate Cancer.Chemotherapy. 2020;65(5-6):125-133. doi: 10.1159/000511505. Epub 2020 Nov 25. Chemotherapy. 2020. PMID: 33238278
-
Inhibition of ABCB1 expression overcomes acquired docetaxel resistance in prostate cancer.Mol Cancer Ther. 2013 Sep;12(9):1829-36. doi: 10.1158/1535-7163.MCT-13-0208. Epub 2013 Jul 16. Mol Cancer Ther. 2013. PMID: 23861346 Free PMC article.
-
Integrative Chinese-Western medicine strategy to overcome docetaxel resistance in prostate cancer.J Ethnopharmacol. 2024 Sep 15;331:118265. doi: 10.1016/j.jep.2024.118265. Epub 2024 Apr 25. J Ethnopharmacol. 2024. PMID: 38677579 Review.
-
Nanoways to overcome docetaxel resistance in prostate cancer.Drug Resist Updat. 2014 Apr;17(1-2):13-23. doi: 10.1016/j.drup.2014.04.001. Epub 2014 Apr 5. Drug Resist Updat. 2014. PMID: 24853766 Free PMC article. Review.
Cited by
-
Targeting the glutamine metabolism to suppress cell proliferation in mesenchymal docetaxel-resistant prostate cancer.Oncogene. 2024 Jun;43(26):2038-2050. doi: 10.1038/s41388-024-03059-4. Epub 2024 May 15. Oncogene. 2024. PMID: 38750263 Free PMC article.
-
Genetic Signatures for Distinguishing Chemo-Sensitive from Chemo-Resistant Responders in Prostate Cancer Patients.Curr Issues Mol Biol. 2024 Mar 11;46(3):2263-2277. doi: 10.3390/cimb46030145. Curr Issues Mol Biol. 2024. PMID: 38534761 Free PMC article.
-
Docetaxel Administration via Novel Hierarchical Nanoparticle Reduces Proinflammatory Cytokine Levels in Prostate Cancer Cells.Cancers (Basel). 2025 May 23;17(11):1758. doi: 10.3390/cancers17111758. Cancers (Basel). 2025. PMID: 40507238 Free PMC article.
-
ABCC1 and ABCC10 as predictive biomarkers of docetaxel treatment response in prostate cancer.Curr Res Pharmacol Drug Discov. 2025 Mar 14;8:100216. doi: 10.1016/j.crphar.2025.100216. eCollection 2025. Curr Res Pharmacol Drug Discov. 2025. PMID: 40213361 Free PMC article.
-
Glutamine Metabolism and Prostate Cancer.Cancers (Basel). 2024 Aug 18;16(16):2871. doi: 10.3390/cancers16162871. Cancers (Basel). 2024. PMID: 39199642 Free PMC article. Review.
References
-
- Cornford P., van den Bergh R.C.N., Briers E., Van den Broeck T., Cumberbatch M.G., De Santis M., Fanti S., Fossati N., Gandaglia G., Gillessen S., et al. EAU-EANM-ESTRO-ESUR-SIOG guidelines on prostate cancer. Part II—2020 update: Treatment of relapsing and metastatic prostate cancer. Eur. Urol. 2021;79:263–282. doi: 10.1016/j.eururo.2020.09.046. - DOI - PubMed
-
- Petrylak D.P., Tangen C.M., Hussain M.H., Lara P.N., Jr., Jones J.A., Taplin M.E., Burch P.A., Berry D., Moinpour C., Kohli M., et al. Docetaxel and estramustine compared with mitoxantrone and prednisone for advanced refractory prostate cancer. N. Engl. J. Med. 2004;351:1513–1520. doi: 10.1056/NEJMoa041318. - DOI - PubMed
-
- Puhr M., Hoefer J., Schafer G., Erb H.H., Oh S.J., Klocker H., Heidegger I., Neuwirt H., Culig Z. Epithelial-to-mesenchymal transition leads to docetaxel resistance in prostate cancer and is mediated by reduced expression of miR-200c and miR-205. Am. J. Pathol. 2012;181:2188–2201. doi: 10.1016/j.ajpath.2012.08.011. - DOI - PubMed
-
- Donix L., Erb H.H.H., Peitzsch C., Dubrovska A., Pfeifer M., Thomas C., Fuessel S., Erdmann K. Acquired resistance to irradiation or docetaxel is not associated with cross-resistance to cisplatin in prostate cancer cell lines. J. Cancer Res. Clin. Oncol. 2022;148:1313–1324. doi: 10.1007/s00432-022-03914-5. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

