Blood Plasma Proteome: A Meta-Analysis of the Results of Protein Quantification in Human Blood by Targeted Mass Spectrometry
- PMID: 36614211
- PMCID: PMC9821253
- DOI: 10.3390/ijms24010769
Blood Plasma Proteome: A Meta-Analysis of the Results of Protein Quantification in Human Blood by Targeted Mass Spectrometry
Abstract
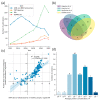
A meta-analysis of the results of targeted quantitative screening of human blood plasma was performed to generate a reference standard kit that can be used for health analytics. The panel included 53 of the 296 proteins that form a “stable” part of the proteome of a healthy individual; these proteins were found in at least 70% of samples and were characterized by an interindividual coefficient of variation <40%. The concentration range of the selected proteins was 10−10−10−3 M and enrichment analysis revealed their association with rare familial diseases. The concentration of ceruloplasmin was reduced by approximately three orders of magnitude in patients with neurological disorders compared to healthy volunteers, and those of gelsolin isoform 1 and complement factor H were abruptly reduced in patients with lung adenocarcinoma. Absolute quantitative data of the individual proteome of a healthy and diseased individual can be used as the basis for personalized medicine and health monitoring. Storage over time allows us to identify individual biomarkers in the molecular landscape and prevent pathological conditions.
Keywords: human proteome project; knowledge databases; proteome; targeted mass spectrometry analysis.
Conflict of interest statement
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Skates S.J., Gillette M.A., LaBaer J., Carr S.A., Anderson L., Liebler D.C., Ransohoff D., Rifai N., Kondratovich M., Težak Ž., et al. Statistical design for biospecimen cohort size in proteomics-based biomarker discovery and verification studies. J. Proteome Res. 2013;12:5383–5394. doi: 10.1021/pr400132j. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

