High-Yield Characterization of Single Molecule Interactions with DeepTipTM Atomic Force Microscopy Probes
- PMID: 36615422
- PMCID: PMC9822271
- DOI: 10.3390/molecules28010226
High-Yield Characterization of Single Molecule Interactions with DeepTipTM Atomic Force Microscopy Probes
Abstract
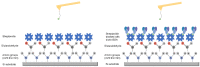
Single molecule interactions between biotin and streptavidin were characterized with functionalized DeepTipTM probes and used as a model system to develop a comprehensive methodology for the high-yield identification and analysis of single molecular events. The procedure comprises the covalent binding of the target molecule to a surface and of the sensing molecule to the DeepTipTM probe, so that the interaction between both chemical species can be characterized by obtaining force-displacement curves in an atomic force microscope. It is shown that molecular resolution is consistently attained with a percentage of successful events higher than 90% of the total number of recorded curves, and a very low level of unspecific interactions. The combination of both features is a clear indication of the robustness and versatility of the proposed methodology.
Keywords: AFM; affinity atomic force microscopy; biotin; functionalization; streptavidin.
Conflict of interest statement
This study was partially funded by the company Bioactive Surfaces S.L. that provided the DeepTipTM probes. DeepTipTM probes are commercially available from the company, and free trial batches of DeepTipTM probes may be available upon request to the company or after contacting the corresponding author.
Figures






Similar articles
-
Statistical Study of Low-Intensity Single-Molecule Recognition Events Using DeepTipTM Probes: Application to the Pru p 3-Phytosphingosine System.Biomimetics (Basel). 2023 Dec 8;8(8):595. doi: 10.3390/biomimetics8080595. Biomimetics (Basel). 2023. PMID: 38132534 Free PMC article.
-
Identification of Individual Target Molecules Using Antibody-Decorated DeepTipTM Atomic-Force Microscopy Probes.Biomimetics (Basel). 2024 Mar 22;9(4):192. doi: 10.3390/biomimetics9040192. Biomimetics (Basel). 2024. PMID: 38667203 Free PMC article.
-
Reconsideration of dynamic force spectroscopy analysis of streptavidin-biotin interactions.Int J Mol Sci. 2010 May 13;11(5):2134-51. doi: 10.3390/ijms11052134. Int J Mol Sci. 2010. PMID: 20559507 Free PMC article.
-
Chemical modifications of atomic force microscopy tips.Methods Mol Biol. 2011;736:457-83. doi: 10.1007/978-1-61779-105-5_28. Methods Mol Biol. 2011. PMID: 21660744 Review.
-
Lab on a tip: Applications of functional atomic force microscopy for the study of electrical properties in biology.Acta Biomater. 2019 Nov;99:33-52. doi: 10.1016/j.actbio.2019.08.023. Epub 2019 Aug 16. Acta Biomater. 2019. PMID: 31425893 Review.
Cited by
-
Statistical Study of Low-Intensity Single-Molecule Recognition Events Using DeepTipTM Probes: Application to the Pru p 3-Phytosphingosine System.Biomimetics (Basel). 2023 Dec 8;8(8):595. doi: 10.3390/biomimetics8080595. Biomimetics (Basel). 2023. PMID: 38132534 Free PMC article.
-
Identification of Individual Target Molecules Using Antibody-Decorated DeepTipTM Atomic-Force Microscopy Probes.Biomimetics (Basel). 2024 Mar 22;9(4):192. doi: 10.3390/biomimetics9040192. Biomimetics (Basel). 2024. PMID: 38667203 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

