Predicting the prognosis in patients with sepsis by a pyroptosis-related gene signature
- PMID: 36618365
- PMCID: PMC9811195
- DOI: 10.3389/fimmu.2022.1110602
Predicting the prognosis in patients with sepsis by a pyroptosis-related gene signature
Abstract
Background: Sepsis remains a life-threatening disease with a high mortality rate that causes millions of deaths worldwide every year. Many studies have suggested that pyroptosis plays an important role in the development and progression of sepsis. However, the potential prognostic and diagnostic value of pyroptosis-related genes in sepsis remains unknown.
Methods: The GSE65682 and GSE95233 datasets were obtained from Gene Expression Omnibus (GEO) database and pyroptosis-related genes were obtained from previous literature and Molecular Signature Database. Univariate cox analysis and least absolute shrinkage and selection operator (LASSO) cox regression analysis were used to select prognostic differentially expressed pyroptosis-related genes and constructed a prognostic risk score. Functional analysis and immune infiltration analysis were used to investigate the biological characteristics and immune cell enrichment in sepsis patients who were classified as low- or high-risk based on their risk score. Then the correlation between pyroptosis-related genes and immune cells was analyzed and the diagnostic value of the selected genes was assessed using the receiver operating characteristic curve.
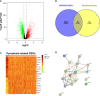
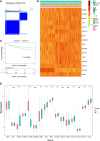
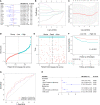
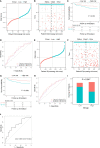
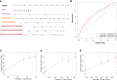
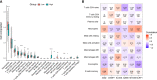
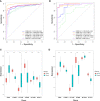
Results: A total of 16 pyroptosis-related differentially expressed genes were identified between sepsis patients and healthy individuals. A six-gene-based (GZMB, CHMP7, NLRP1, MYD88, ELANE, and AIM2) prognostic risk score was developed. Based on the risk score, sepsis patients were divided into low- and high-risk groups, and patients in the low-risk group had a better prognosis. Functional enrichment analysis found that NOD-like receptor signaling pathway, hematopoietic cell lineage, and other immune-related pathways were enriched. Immune infiltration analysis showed that some innate and adaptive immune cells were significantly different between low- and high-risk groups, and correlation analysis revealed that all six genes were significantly correlated with neutrophils. Four out of six genes (GZMB, CHMP7, NLRP1, and AIM2) also have potential diagnostic value in sepsis diagnosis.
Conclusion: We developed and validated a novel prognostic predictive risk score for sepsis based on six pyroptosis-related genes. Four out of the six genes also have potential diagnostic value in sepsis diagnosis.
Keywords: diagnosis; gene; prediction; prognosis; pyroptosis; sepsis.
Copyright © 2022 Liang, Xing, Chen, Peng, Song and Zou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Deciphering the role of pyroptosis-related genes and natural killer T cells in sepsis pathogenesis: a comprehensive bioinformatics and Mendelian randomization analysis.J Physiol Pharmacol. 2025 Apr;76(2). doi: 10.26402/jpp.2025.2.10. Epub 2025 May 5. J Physiol Pharmacol. 2025. PMID: 40350654
-
Exploring the role of pyroptosis and immune infiltration in sepsis based on bioinformatic analysis.Immunobiology. 2024 Sep;229(5):152826. doi: 10.1016/j.imbio.2024.152826. Epub 2024 Jun 10. Immunobiology. 2024. PMID: 38981197
-
A novel pyroptosis-related gene signature for predicting laryngeal carcinoma prognosis.Int J Clin Exp Pathol. 2022 Aug 15;15(8):301-315. eCollection 2022. Int J Clin Exp Pathol. 2022. PMID: 36106070 Free PMC article.
-
System analysis based on the pyroptosis-related genes identifies GSDMC as a novel therapy target for pancreatic adenocarcinoma.J Transl Med. 2022 Oct 5;20(1):455. doi: 10.1186/s12967-022-03632-z. J Transl Med. 2022. PMID: 36199146 Free PMC article.
-
Role of toll-like receptor-mediated pyroptosis in sepsis-induced cardiomyopathy.Biomed Pharmacother. 2023 Nov;167:115493. doi: 10.1016/j.biopha.2023.115493. Epub 2023 Sep 19. Biomed Pharmacother. 2023. PMID: 37734261 Review.
Cited by
-
LncRNA SNHG16 promotes LPS-induced human bronchial epithelial cell pyroptosis through miR-339-5p/NLRP1 axis mediation.Cent Eur J Immunol. 2024;49(4):345-365. doi: 10.5114/ceji.2024.145876. Epub 2024 Dec 12. Cent Eur J Immunol. 2024. PMID: 39944259 Free PMC article.
-
CircIRAK3 Promotes Neutrophil Extracellular Trap Formation by Improving the Stability of ELANE mRNA in Sepsis.Inflammation. 2025 Aug;48(4):2503-2515. doi: 10.1007/s10753-024-02206-z. Epub 2024 Dec 21. Inflammation. 2025. PMID: 39707013 Free PMC article.
-
Exploring the Role of Different Cell-Death-Related Genes in Sepsis Diagnosis Using a Machine Learning Algorithm.Int J Mol Sci. 2023 Sep 29;24(19):14720. doi: 10.3390/ijms241914720. Int J Mol Sci. 2023. PMID: 37834169 Free PMC article.
-
Immune-associated molecular classification and prognosis signature of sepsis.PLoS One. 2025 Jun 12;20(6):e0326083. doi: 10.1371/journal.pone.0326083. eCollection 2025. PLoS One. 2025. PMID: 40504881 Free PMC article.
-
Analysis of Immune and Prognostic-Related lncRNA PRKCQ-AS1 for Predicting Prognosis and Regulating Effect in Sepsis.J Inflamm Res. 2024 Jan 12;17:279-299. doi: 10.2147/JIR.S433057. eCollection 2024. J Inflamm Res. 2024. PMID: 38229689 Free PMC article.
References
-
- Fleischmann-Struzek C, Mellhammar L, Rose N, Cassini A, Rudd KE, Schlattmann P, et al. . Incidence and mortality of hospital- and icu-treated sepsis: Results from an updated and expanded systematic review and meta-analysis. Intensive Care Med (2020) 46(8):1552–62. doi: 10.1007/s00134-020-06151-x - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

