Chicken CSF2 and IL-4-, and CSF2-dependent bone marrow cultures differentiate into macrophages over time
- PMID: 36618373
- PMCID: PMC9812659
- DOI: 10.3389/fimmu.2022.1064084
Chicken CSF2 and IL-4-, and CSF2-dependent bone marrow cultures differentiate into macrophages over time
Abstract
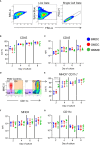
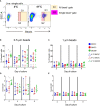
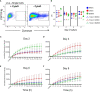
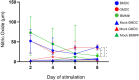
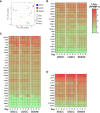
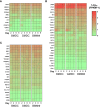
Chicken bone marrow-derived macrophages (BMMΦ) and dendritic cells (BMDC) are utilized as models to study the mononuclear phagocytic system (MPS). A widely used method to generate macrophages and DC in vitro is to culture bone marrow cells in the presence of colony-stimulating factor-1 (CSF1) to differentiate BMMΦ and granulocyte-macrophage-CSF (GM-CSF, CSF2) and interleukin-4 (IL-4) to differentiate BMDC, while CSF2 alone can lead to the development of granulocyte-macrophage-CSF-derived DC (GMDC). However, in chickens, the MPS cell lineages and their functions represented by these cultures are poorly understood. Here, we decipher the phenotypical, functional and transcriptional differences between chicken BMMΦ and BMDC along with examining differences in DC cultures grown in the absence of IL-4 on days 2, 4, 6 and 8 of culture. BMMΦ cultures develop into a morphologically homogenous cell population in contrast to the BMDC and GMDC cultures, which produce morphologically heterogeneous cell cultures. At a phenotypical level, all cultures contained similar cell percentages and expression levels of MHCII, CD11c and CSF1R-transgene, whilst MRC1L-B expression decreased over time in BMMΦ. All cultures were efficiently able to uptake 0.5 µm beads, but poorly phagocytosed 1 µm beads. Little difference was observed in the kinetics of phagosomal acidification across the cultures on each day of analysis. Temporal transcriptomic analysis indicated that all cultures expressed high levels of CSF3R, MERTK, SEPP1, SPI1 and TLR4, genes associated with macrophages in mammals. In contrast, low levels of FLT3, XCR1 and CAMD1, genes associated with DC, were expressed at day 2 in BMDC and GMDC after which expression levels decreased. Collectively, chicken CSF2 + IL-4- and CSF2-dependent BM cultures represent cells of the macrophage lineage rather than inducing conventional DC.
Keywords: bone marrow cell cultures; chicken; dendritic cell; macrophage; transcriptome.
Copyright © 2022 Borowska, Sives, Vervelde and Sutton.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Lardon F, Snoeck HW, Berneman ZN, Van Tendeloo VF, Nijs G, Lenjou M, et al. . Generation of dendritic cells from bone marrow progenitors using gm-csf, tnf-alpha, and additional cytokines: Antagonistic effects of il-4 and ifn-gamma and selective involvement of tnf-alpha receptor-1. Immunology (1997) 91(4):553–9. doi: 10.1046/j.1365-2567.1997.00295.x - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/D/20320000/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M003094/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/10002071/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/D/20002174/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

