Gibberellic acid sensitive dwarf encodes an ARPC2 subunit that mediates gibberellic acid biosynthesis, effects to grain yield in rice
- PMID: 36618614
- PMCID: PMC9813395
- DOI: 10.3389/fpls.2022.1027688
Gibberellic acid sensitive dwarf encodes an ARPC2 subunit that mediates gibberellic acid biosynthesis, effects to grain yield in rice
Abstract
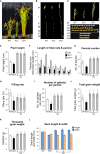
The plant hormone gibberellic acid (GA) is important for plant growth and productivity. Actin-related proteins (ARPs) also play central roles in plant growth, including cell elongation and development. However, the relationships between ARPs and GA signaling and biosynthesis are not fully understood. Here, we isolated OsGASD, encoding an ARP subunit from rice (Oryza sativa), using the Ac/Ds knockout system. The osgasd knockout (Ko) mutation reduced GA3 content in shoots as well as plant growth and height. However, GA application restored the plant height of the osgasd Ko mutant to a height similar to that of the wild type (WT). Rice plants overexpressing OsGASD (Ox) showed increased plant height and grain yield compared to the WT. Transcriptome analysis of flag leaves of OsGASD Ox and osgasd Ko plants revealed that OsGASD regulates cell development and the expression of elongation-related genes. These observations suggest that OsGASD is involved in maintaining GA homeostasis to regulate plant development, thereby affecting rice growth and productivity.
Keywords: Oryza sativa; actin-related protein; gibberellic acid; grain yield; rice growth.
Copyright © 2022 Um, Hong, Han, Jung, Moon, Choi, Basnet, Chung, Lee, Yang and Kim.
Conflict of interest statement
Author BC was employed by NBIT Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Barboza L., Effgen S., Alonso-Blanco C., Kooke R., Keurentjes J. J. B., Koornneef M., et al. . (2013). Arabidopsis semidwarfs evolved from independent mutations in GA20ox1, ortholog to green revolution dwarf alleles in rice and barley. Proc. Natl. Acad. Sci. U. S. A. 110, 15818–15823. doi: 10.1073/pnas.1314979110 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

