SiMeEx, a simplified method for metabolite extraction of adherent mammalian cells
- PMID: 36619169
- PMCID: PMC9812552
- DOI: 10.3389/fmolb.2022.1084060
SiMeEx, a simplified method for metabolite extraction of adherent mammalian cells
Abstract
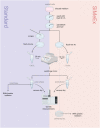
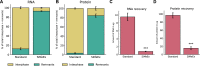
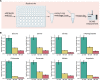
A reliable method for metabolite extraction is central to mass spectrometry-based metabolomics. However, existing methods are lengthy, mostly due to the step of scraping cells from cell culture vessels, which restricts metabolomics in broader application such as lower cell numbers and high-throughput studies. Here, we present a simplified metabolite extraction (SiMeEx) method, to efficiently and quickly extract metabolites from adherent mammalian cells. Our method excludes the cell scraping step and therefore allows for a more efficient extraction of polar metabolites in less than 30 min per 12-well plate. We demonstrate that SiMeEx achieves the same metabolite recovery as using a standard method containing a scraping step, in various immortalized and primary cells. Omitting cell scraping does not compromise the performance of non-targeted and targeted GC-MS analysis, but enables metabolome analysis of cell culture on smaller well sizes down to 96-well plates. Therefore, SiMeEx demonstrates advantages not only on time and resources, but also on the applicability in high-throughput studies.
Keywords: GC-MS; mammalian cells; metabolite extraction; metabolomics; stable isotope labeling.
Copyright © 2022 Henne, Vigh, Märtens, Nonnenmacher, Ohm, Hosseini, More, Lauterbach, Garritsen, Korte, He and Hiller.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Allison D. (2017). Global metabolomics. Nat. Methods 14, 32. 10.1038/nmeth.4112 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

