Identifying biomarkers of differential chemotherapy response in TNBC patient-derived xenografts with a CTD/WGCNA approach
- PMID: 36619972
- PMCID: PMC9813793
- DOI: 10.1016/j.isci.2022.105799
Identifying biomarkers of differential chemotherapy response in TNBC patient-derived xenografts with a CTD/WGCNA approach
Abstract
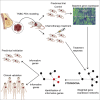
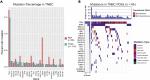
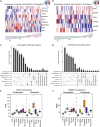
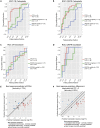
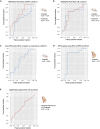
Although systemic chemotherapy remains the standard of care for TNBC, even combination chemotherapy is often ineffective. The identification of biomarkers for differential chemotherapy response would allow for the selection of responsive patients, thus maximizing efficacy and minimizing toxicities. Here, we leverage TNBC PDXs to identify biomarkers of response. To demonstrate their ability to function as a preclinical cohort, PDXs were characterized using DNA sequencing, transcriptomics, and proteomics to show consistency with clinical samples. We then developed a network-based approach (CTD/WGCNA) to identify biomarkers of response to carboplatin (MSI1, TMSB15A, ARHGDIB, GGT1, SV2A, SEC14L2, SERPINI1, ADAMTS20, DGKQ) and docetaxel (c, MAGED4, CERS1, ST8SIA2, KIF24, PARPBP). CTD/WGCNA multigene biomarkers are predictive in PDX datasets (RNAseq and Affymetrix) for both taxane- (docetaxel or paclitaxel) and platinum-based (carboplatin or cisplatin) response, thereby demonstrating cross-expression platform and cross-drug class robustness. These biomarkers were also predictive in clinical datasets, thus demonstrating translational potential.
Keywords: Cancer; Immune response; Omics.
© 2022.
Conflict of interest statement
M.T.L is a Founder of, and an uncompensated Limited Partner in, StemMed Ltd., and an uncompensated Manager in StemMed Holdings L.L.C., its General Partner. MTL is also a Founder of, and equity stake holder in, Tvardi Therapeutics Inc. L.E.D. is a compensated employee of StemMed Ltd. Selected BCM PDX models described herein are exclusively licensed to StemMed Ltd., resulting in tangible property royalties to M.T.L. and L.E.D. University of Utah may license the HCI PDX models described herein to for-profit companies, which may result in tangible property royalties to A.L.W. and B.E.W. Washington University has licensed selected PDX to Envigo, which results in tangible property royalties to S.L. He also received research funding from Pfizer, Takeda Oncology, and Zenopharm, independent of this project. S.L. has received license fees from Envigo. He received research funding from Pfizer, Takeda Oncology, and Zenopharm, outside of this project. O.S, is a compensated employee of, and equity stake holder in, Bluebird Bio. M.J.E. received consulting fees from Abbvie, Sermonix, Pfizer, AstraZeneca, Celgene, NanoString, Puma, Veracyte, Eli Lilly, and Novartis, and he is an equity stockholder and Board of Directors member of BioClassifier. M.J.E. is an inventor on a patent for the Breast Cancer PAM50-based assay, Prosigna, which is marketed by Veracyte. M.J.E. also receives royalties from Washington University in St Louis when the WHIM PDX lines are licensed to for-profit companies.
Figures



References
-
- Garrido-Castro A.C., Lin N.U., Polyak K. Insights into molecular classifications of triple-negative breast cancer: improving patient selection for treatment. Cancer Discov. 2019;9:176–198. doi: 10.1158/2159-8290.cd-18-1177. - DOI - PMC - PubMed
-
- Cocco S., Piezzo M., Calabrese A., Cianniello D., Caputo R., Lauro V.D., Fusco G., Gioia G.d., De Laurentiis M., Licenziato M., Laurentiis M. de. Biomarkers in triple-negative breast cancer: state-of-the-art and future perspectives. Int. J. Mol. Sci. 2020;21:4579. doi: 10.3390/ijms21134579. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

