Motility increase of adherent invasive Escherichia coli (AIEC) induced by a sub-inhibitory concentration of recombinant endolysin LysPA90
- PMID: 36619993
- PMCID: PMC9814724
- DOI: 10.3389/fmicb.2022.1093670
Motility increase of adherent invasive Escherichia coli (AIEC) induced by a sub-inhibitory concentration of recombinant endolysin LysPA90
Erratum in
-
Corrigendum: Motility increase of Adherent Invasive Escherichia coli (AIEC) induced by a sub-inhibitory concentration of recombinant endolysin LysPA90.Front Microbiol. 2023 Aug 15;14:1272581. doi: 10.3389/fmicb.2023.1272581. eCollection 2023. Front Microbiol. 2023. PMID: 37664130 Free PMC article.
Abstract
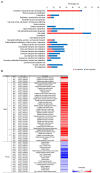
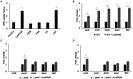
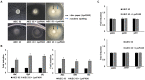
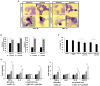
Endolysins are bacteriophage enzymes required for the eruption of phages from inside host bacteria via the degradation of the peptidoglycan cell wall. Recombinant endolysins are increasingly being seen as potential antibacterial candidates, with a number currently undergoing clinical trials. Bacteriophage PBPA90 infecting Pseudomonas aeruginosa harbors a gene encoding an endolysin, lysPA90. Herein, recombinant LysPA90 demonstrated an intrinsic antibacterial activity against Escherichia coli in vitro. It was observed that a sub-inhibitory concentration of the recombinant protein induced the upregulation of genes related to flagella biosynthesis in a commensal E. coli strain. Increases in the number of bacterial flagella, and in motility, were experimentally substantiated. The treatment caused membrane stress, leading to the upregulation of genes rpoE, rpoH, dnaK, dnaJ, and flhC, which are upstream regulators of flagella biosynthesis. When adherent invasive Escherichia coli (AIEC) strains were treated with subinhibitory concentrations of the endolysin, bacterial adhesion and invasion into intestinal epithelial Caco-2 cells was seen to visibly increase under microscopic examination. Bacterial counting further corroborated this adhesion and invasion of AIEC strains into Caco-2 cells, with a resultant slight decrease in the viability of Caco-2 cells then being observed. Additionally, genes related to flagella expression were also upregulated in the AIEC strains. Finally, the enhanced expression of the proinflammatory cytokine genes TNF-α, IL-6, IL-8, and MCP1 in Caco-2 cells was noted after the increased invasion of the AIEC strains. While novel treatments involving endolysins offer great promise, these results highlight the need for the further exploration of possible unanticipated and unintended effects.
Keywords: AIEC; bacteriophage; endolysin; flagella; membrane stress.
Copyright © 2022 Hwang, Jo, Kim, Yoon, Hong, Kim and Myung.
Conflict of interest statement
YH, HH, MK, and HM are employed by LyseNTech. Co. Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Corrigendum: Motility increase of Adherent Invasive Escherichia coli (AIEC) induced by a sub-inhibitory concentration of recombinant endolysin LysPA90.Front Microbiol. 2023 Aug 15;14:1272581. doi: 10.3389/fmicb.2023.1272581. eCollection 2023. Front Microbiol. 2023. PMID: 37664130 Free PMC article.
-
Adaptation of adherent-invasive E. coli to gut environment: Impact on flagellum expression and bacterial colonization ability.Gut Microbes. 2020 May 3;11(3):364-380. doi: 10.1080/19490976.2017.1421886. Epub 2018 Mar 1. Gut Microbes. 2020. PMID: 29494278 Free PMC article.
-
Development of Heptylmannoside-Based Glycoconjugate Antiadhesive Compounds against Adherent-Invasive Escherichia coli Bacteria Associated with Crohn's Disease.mBio. 2015 Nov 17;6(6):e01298-15. doi: 10.1128/mBio.01298-15. mBio. 2015. PMID: 26578673 Free PMC article.
-
Pathogenesis of adherent-invasive Escherichia coli.Future Microbiol. 2013 Oct;8(10):1289-300. doi: 10.2217/fmb.13.94. Future Microbiol. 2013. PMID: 24059919 Review.
-
Escherichia coli in chronic inflammatory bowel diseases: An update on adherent invasive Escherichia coli pathogenicity.World J Gastrointest Pathophysiol. 2014 Aug 15;5(3):213-27. doi: 10.4291/wjgp.v5.i3.213. World J Gastrointest Pathophysiol. 2014. PMID: 25133024 Free PMC article. Review.
Cited by
-
Phage Endolysins as Promising and Effective Candidates for Use Against Uropathogenic Escherichia coli.Viruses. 2025 Apr 13;17(4):560. doi: 10.3390/v17040560. Viruses. 2025. PMID: 40285003 Free PMC article. Review.
References
-
- Barker C. S., Prüss B. M. (2005). FlhD/FlhC, a global transcriptional regulator in Escherichia coli,” in Global Regulatory Networks in Enteric Bacteria. Research Signpost, Trivandrum, Kerala, India, pp 13–30.
LinkOut - more resources
Full Text Sources

