Genetic characterization and molecular epidemiology of Coxsackievirus A12 from mainland China during 2010-2019
- PMID: 36620057
- PMCID: PMC9811122
- DOI: 10.3389/fmicb.2022.988538
Genetic characterization and molecular epidemiology of Coxsackievirus A12 from mainland China during 2010-2019
Abstract
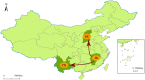
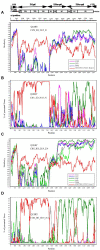
Coxsackievirus A12 (CVA12) is an enterovirus that has been isolated in many countries in recent years. However, studies on CVA12 are limited, and its effective population size, evolutionary dynamics and recombination patterns have not been clarified now. In this study, we described the phylogenetic characteristics of 16 CVA12 strains isolated from pediatric HFMD patients in mainland China from 2010 to 2019. Comparison of the nucleotide sequences and amino acid sequences with the CVA12 prototype strain revealed that the 16 CVA12 strains are identical in 78.8-79% and 94-94.2%, respectively. A phylodynamic analysis based on the 16 full-length VP1 sequences from this study and 21 sequences obtained from GenBank revealed a mean substitution rate of 6.61 × 10-3 substitutions/site/year (95% HPD: 5.16-8.20 × 10-3), dating the time to most recent common ancestor (tMRCA) of CVA12 back to 1946 (95% HPD: 1942-1947). The Bayesian skyline plot showed that the effective population size has experienced twice dynamic fluctuations since 2007. Phylogeographic analysis identified two significant migration pathways, indicating the existence of cross-provincial transmission of CVA12 in mainland China. Recombination analysis revealed two recombination patterns between 16 CVA12 strains and other EV-A, suggesting that there may be extensive genetic exchange between CVA12 and other enteroviruses. In summary, a total of 16 full-length CVA12 strains were reported in this study, providing valuable references for further studies of CVA12 worldwide.
Keywords: Coxsackievirus A12; enteroviruses; evolutionary dynamics; foot and mouth disease; hand; phylogenetic analysis; recombination.
Copyright © 2022 Guo, Zhao, Zhang, Wang, Yu, Tan, Lu, Xiao, Ji, Zhu, Wang, Yang, Han, Xu and Yan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Molecular Epidemiology and Evolution of Coxsackievirus A14.Viruses. 2023 Nov 26;15(12):2323. doi: 10.3390/v15122323. Viruses. 2023. PMID: 38140564 Free PMC article.
-
Molecular characteristics of a coxsackievirus A12 strain in Zhejiang of China, 2019.Virol J. 2022 Oct 12;19(1):160. doi: 10.1186/s12985-022-01892-1. Virol J. 2022. PMID: 36224635 Free PMC article.
-
Genetic characterization of emerging coxsackievirus A12 associated with hand, foot and mouth disease in Qingdao, China.Arch Virol. 2014 Sep;159(9):2497-502. doi: 10.1007/s00705-014-2067-6. Epub 2014 May 6. Arch Virol. 2014. PMID: 24796551
-
Genomic surveillance reveals low-level circulation of two subtypes of genogroup C coxsackievirus A10 in Nanchang, Jiangxi Province, China, 2015-2023.Front Microbiol. 2024 Sep 17;15:1459917. doi: 10.3389/fmicb.2024.1459917. eCollection 2024. Front Microbiol. 2024. PMID: 39355427 Free PMC article.
-
Molecular Epidemiology and Evolution of Coxsackievirus A9.Viruses. 2022 Apr 15;14(4):822. doi: 10.3390/v14040822. Viruses. 2022. PMID: 35458552 Free PMC article.
References
-
- Chen S., Huang Y., Li W., Chiu C.-H., Huang C.-G., Tsao K.-C., et al. (2010). Comparison of clinical features between coxsackievirus A2 and enterovirus 71 during the enterovirus outbreak in Taiwan, 2008: a children's hospital experience. J. Microbiol. Immunol. Infect. 43, 99–104. doi: 10.1016/S1684-1182(10)60016-3, PMID: - DOI - PubMed
LinkOut - more resources
Full Text Sources

