Graded impact of obstacle size on scanning by RNase E
- PMID: 36620905
- PMCID: PMC9943677
- DOI: 10.1093/nar/gkac1242
Graded impact of obstacle size on scanning by RNase E
Abstract
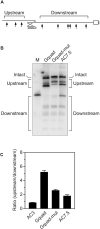
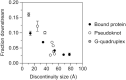
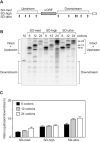
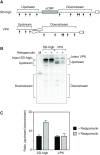
In countless bacterial species, the lifetimes of most mRNAs are controlled by the regulatory endonuclease RNase E, which preferentially degrades RNAs bearing a 5' monophosphate and locates cleavage sites within them by scanning linearly from the 5' terminus along single-stranded regions. Consequently, its rate of cleavage at distal sites is governed by any obstacles that it may encounter along the way, such as bound proteins or ribosomes or base pairing that is coaxial with the path traversed by this enzyme. Here, we report that the protection afforded by such obstacles is dependent on the size and persistence of the structural discontinuities they create, whereas the molecular composition of obstacles to scanning is of comparatively little consequence. Over a broad range of sizes, incrementally larger discontinuities are incrementally more protective, with corresponding effects on mRNA stability. The graded impact of such obstacles suggests possible explanations for why their effect on scanning is not an all-or-none phenomenon dependent simply on whether the size of the resulting discontinuity exceeds the step length of RNase E.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
Obstacles to Scanning by RNase E Govern Bacterial mRNA Lifetimes by Hindering Access to Distal Cleavage Sites.Mol Cell. 2019 Apr 18;74(2):284-295.e5. doi: 10.1016/j.molcel.2019.01.044. Epub 2019 Mar 6. Mol Cell. 2019. PMID: 30852060 Free PMC article.
-
In vitro analysis of mRNA processing by RNase E in the pap operon of Escherichia coli.Mol Microbiol. 1996 Jul;21(1):55-68. doi: 10.1046/j.1365-2958.1996.6121101.x. Mol Microbiol. 1996. PMID: 8843434
-
Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB.Nucleic Acids Res. 2005 Mar 21;33(5):1678-89. doi: 10.1093/nar/gki313. Print 2005. Nucleic Acids Res. 2005. PMID: 15781494 Free PMC article.
-
Translation initiation and the fate of bacterial mRNAs.FEMS Microbiol Rev. 2006 Nov;30(6):967-79. doi: 10.1111/j.1574-6976.2006.00043.x. Epub 2006 Sep 21. FEMS Microbiol Rev. 2006. PMID: 16989654 Review.
-
Killer and protective ribosomes.Prog Mol Biol Transl Sci. 2009;85:423-66. doi: 10.1016/S0079-6603(08)00811-8. Prog Mol Biol Transl Sci. 2009. PMID: 19215779 Review.
Cited by
-
Improved RNA stability estimation through Bayesian modeling reveals most Salmonella transcripts have subminute half-lives.Proc Natl Acad Sci U S A. 2024 Apr 2;121(14):e2308814121. doi: 10.1073/pnas.2308814121. Epub 2024 Mar 25. Proc Natl Acad Sci U S A. 2024. PMID: 38527194 Free PMC article.
-
The membrane-targeting-sequence motif is required for exhibition of recessive resurrection in Escherichia coli RNase E.Nucleic Acids Res. 2025 Jan 24;53(3):gkaf055. doi: 10.1093/nar/gkaf055. Nucleic Acids Res. 2025. PMID: 39898549 Free PMC article.
-
RNase E searches for cleavage sites in RNA by linear diffusion: direct evidence from single-molecule FRET.Nucleic Acids Res. 2024 Jun 24;52(11):6674-6686. doi: 10.1093/nar/gkae279. Nucleic Acids Res. 2024. PMID: 38647084 Free PMC article.
-
The small non-coding RNA B11 regulates multiple facets of Mycobacterium abscessus virulence.PLoS Pathog. 2023 Aug 21;19(8):e1011575. doi: 10.1371/journal.ppat.1011575. eCollection 2023 Aug. PLoS Pathog. 2023. PMID: 37603560 Free PMC article.
References
-
- Mackie G.A. RNase E: at the interface of bacterial RNA processing and decay. Nat. Rev. Microbiol. 2013; 11:45–57. - PubMed
-
- McDowall K.J., Lin-Chao S., Cohen S.N.. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J. Biol. Chem. 1994; 269:10790–10796. - PubMed
-
- Lin-Chao S., Wong T.-T., McDowall K.J., Cohen S.N.. Effects of nucleotide sequence on the specificity of rne-dependent and RNase E-mediated cleavages of RNA I encoded by the pBR322 plasmid. J. Biol. Chem. 1994; 269:10797–10803. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

