Natural and synthetic 2-oxoglutarate derivatives are substrates for oncogenic variants of human isocitrate dehydrogenase 1 and 2
- PMID: 36621625
- PMCID: PMC9939733
- DOI: 10.1016/j.jbc.2023.102873
Natural and synthetic 2-oxoglutarate derivatives are substrates for oncogenic variants of human isocitrate dehydrogenase 1 and 2
Abstract
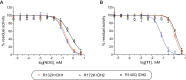
Variants of isocitrate dehydrogenase (IDH) 1 and 2 (IDH1/2) alter metabolism in cancer cells by catalyzing the NADPH-dependent reduction of 2-oxoglutarate (2OG) to (2R)-hydroxyglutarate. However, it is unclear how derivatives of 2OG can affect cancer cell metabolism. Here, we used synthetic C3- and C4-alkylated 2OG derivatives to investigate the substrate selectivities of the most common cancer-associated IDH1 variant (R132H IDH1), of two cancer-associated IDH2 variants (R172K IDH2, R140Q IDH2), and of WT IDH1/2. Absorbance-based, NMR, and electrochemical assays were employed to monitor WT IDH1/2 and IDH1/2 variant-catalyzed 2OG derivative turnover in the presence and absence of 2OG. Our results reveal that 2OG derivatives can serve as substrates of the investigated IDH1/2 variants, but not of WT IDH1/2, and have the potential to act as 2OG-competitive inhibitors. Kinetic parameters reveal that some 2OG derivatives, including the natural product 3-methyl-2OG, are equally or even more efficient IDH1/2 variant substrates than 2OG. Furthermore, NMR and mass spectrometry studies confirmed IDH1/2 variant-catalyzed production of alcohols in the cases of the 3-methyl-, 3-butyl-, and 3-benzyl-substituted 2OG derivatives; a crystal structure of 3-butyl-2OG with an IDH1 variant (R132C/S280F IDH1) reveals active site binding. The combined results highlight the potential for (i) IDH1/2 variant-catalyzed reduction of 2-oxoacids other than 2OG in cells, (ii) modulation of IDH1/2 variant activity by 2-oxoacid natural products, including some present in common foods, (iii) inhibition of IDH1/2 variants via active site binding rather than the established allosteric mode of inhibition, and (iv) possible use of IDH1/2 variants as biocatalysts.
Keywords: (R)-2-hydroxyglutarate; 2-oxoacids; 2-oxoglutarate; 2HG; 2OG; IDH; acute myeloid leukemia; alternative substrates; cancer metabolism-related IDH mutations; epigenetics; isocitrate dehydrogenase; α-ketoacids; α-ketoglutarate.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflict of interest with the contents of this article.
Figures





References
-
- Xu X., Zhao J., Xu Z., Peng B., Huang Q., Arnold E., et al. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J. Biol. Chem. 2004;279:33946–33957. - PubMed
-
- Dang L., Su S.-S.M. Isocitrate dehydrogenase mutation and (R)-2-hydroxyglutarate: from basic discovery to therapeutics development. Annu. Rev. Biochem. 2017;86:305–331. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

