Repeat-associated non-AUG translation induces cytoplasmic aggregation of CAG repeat-containing RNAs
- PMID: 36623192
- PMCID: PMC9934169
- DOI: 10.1073/pnas.2215071120
Repeat-associated non-AUG translation induces cytoplasmic aggregation of CAG repeat-containing RNAs
Abstract
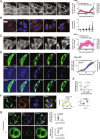
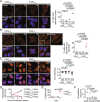
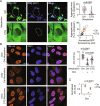
CAG trinucleotide repeat expansions cause several neurodegenerative diseases, including Huntington's disease and spinocerebellar ataxia. RNAs with expanded CAG repeats contribute to disease in two unusual ways. First, these repeat-containing RNAs may agglomerate in the nucleus as foci that sequester several RNA-binding proteins. Second, these RNAs may undergo aberrant repeat-associated non-AUG (RAN) translation in multiple frames and produce aggregation-prone proteins. The relationship between RAN translation and RNA foci, and their relative contributions to cellular dysfunction, are unclear. Here, we show that CAG repeat-containing RNAs that undergo RAN translation first accumulate at nuclear foci and, over time, are exported to the cytoplasm. In the cytoplasm, these RNAs are initially dispersed but, upon RAN translation, aggregate with the RAN translation products. These RNA-RAN protein agglomerates sequester various RNA-binding proteins and are associated with the disruption of nucleocytoplasmic transport and cell death. In contrast, RNA accumulation at nuclear foci alone does not produce discernable defects in nucleocytoplasmic transport or cell viability. Inhibition of RAN translation prevents cytoplasmic RNA aggregation and alleviates cell toxicity. Our findings demonstrate that RAN translation-induced RNA-protein aggregation correlates with the key pathological hallmarks observed in disease and suggest that cytoplasmic RNA aggregation may be an underappreciated phenomenon in CAG trinucleotide repeat expansion disorders.
Keywords: RAN translation; RNA aggregation; RNA localization; Repeat expansion diseases.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Jiang H., Mankodi A., Swanson M. S., Moxley R. T., Thornton C. A., Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum. Mol. Genet. 13, 3079–3088 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

