Tumor-intrinsic IRE1α signaling controls protective immunity in lung cancer
- PMID: 36624093
- PMCID: PMC9829901
- DOI: 10.1038/s41467-022-35584-9
Tumor-intrinsic IRE1α signaling controls protective immunity in lung cancer
Abstract
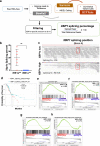
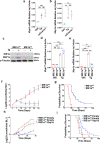
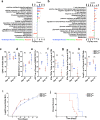
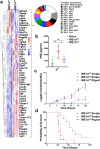
IRE1α-XBP1 signaling is emerging as a central orchestrator of malignant progression and immunosuppression in various cancer types. Employing a computational XBP1s detection method applied to TCGA datasets, we demonstrate that expression of the XBP1s mRNA isoform predicts poor survival in non-small cell lung cancer (NSCLC) patients. Ablation of IRE1α in malignant cells delays tumor progression and extends survival in mouse models of NSCLC. This protective effect is accompanied by alterations in intratumoral immune cell subsets eliciting durable adaptive anti-cancer immunity. Mechanistically, cancer cell-intrinsic IRE1α activation sustains mPGES-1 expression, enabling production of the immunosuppressive lipid mediator prostaglandin E2. Accordingly, restoring mPGES-1 expression in IRE1αKO cancer cells rescues normal tumor progression. We have developed an IRE1α gene signature that predicts immune cell infiltration and overall survival in human NSCLC. Our study unveils an immunoregulatory role for cancer cell-intrinsic IRE1α activation and suggests that targeting this pathway may help enhance anti-tumor immunity in NSCLC.
© 2023. The Author(s).
Conflict of interest statement
J.R.C.-R. is a scientific consultant for NextRNA Therapeutics, Inc. and Autoimmunity Biologic Solutions, Inc, and holds patents on IRE1α modulation for the treatment of disease. O.E. is supported by Janssen, J&J, Astra-Zeneca, Volastra, and Eli Lilly research grants. He is a scientific advisor and equity holder in Freenome, Owkin, Volastra Therapeutics, and One Three Biotech and a paid scientific advisor to Champions Oncology and Pionyr Immunotherapeutics. The remaining authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

