SARS-CoV-2 diagnostic testing rates determine the sensitivity of genomic surveillance programs
- PMID: 36624344
- PMCID: PMC9839449
- DOI: 10.1038/s41588-022-01267-w
SARS-CoV-2 diagnostic testing rates determine the sensitivity of genomic surveillance programs
Abstract
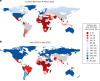
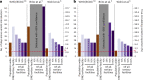
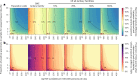
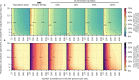
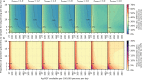
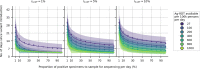
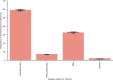
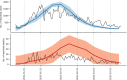
The first step in SARS-CoV-2 genomic surveillance is testing to identify people who are infected. However, global testing rates are falling as we emerge from the acute health emergency and remain low in many low- and middle-income countries (mean = 27 tests per 100,000 people per day). We simulated COVID-19 epidemics in a prototypical low- and middle-income country to investigate how testing rates, sampling strategies and sequencing proportions jointly impact surveillance outcomes, and showed that low testing rates and spatiotemporal biases delay time to detection of new variants by weeks to months and can lead to unreliable estimates of variant prevalence, even when the proportion of samples sequenced is increased. Accordingly, investments in wider access to diagnostics to support testing rates of approximately 100 tests per 100,000 people per day could enable more timely detection of new variants and reliable estimates of variant prevalence. The performance of global SARS-CoV-2 genomic surveillance programs is fundamentally limited by access to diagnostic testing.
© 2023. The Author(s).
Conflict of interest statement
A.T., E.H., S.C., B.R. and B.E.N. declare that they are employed by FIND, the global alliance for diagnostics. All remaining authors declare no competing interests.
Figures





Update of
-
SARS-CoV-2 diagnostic testing rates determine the sensitivity of genomic surveillance programs.medRxiv [Preprint]. 2022 Sep 16:2022.05.20.22275319. doi: 10.1101/2022.05.20.22275319. medRxiv. 2022. Update in: Nat Genet. 2023 Jan;55(1):26-33. doi: 10.1038/s41588-022-01267-w. PMID: 35664998 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

