Pharos 2023: an integrated resource for the understudied human proteome
- PMID: 36624666
- PMCID: PMC9825581
- DOI: 10.1093/nar/gkac1033
Pharos 2023: an integrated resource for the understudied human proteome
Erratum in
-
Correction to 'Pharos 2023: An integrated resource for the understudied human proteome'.Nucleic Acids Res. 2023 Feb 28;51(4):1999. doi: 10.1093/nar/gkad069. Nucleic Acids Res. 2023. PMID: 36744505 Free PMC article. No abstract available.
Abstract
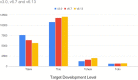
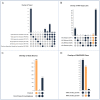
The Illuminating the Druggable Genome (IDG) project aims to improve our understanding of understudied proteins and our ability to study them in the context of disease biology by perturbing them with small molecules, biologics, or other therapeutic modalities. Two main products from the IDG effort are the Target Central Resource Database (TCRD) (http://juniper.health.unm.edu/tcrd/), which curates and aggregates information, and Pharos (https://pharos.nih.gov/), a web interface for fusers to extract and visualize data from TCRD. Since the 2021 release, TCRD/Pharos has focused on developing visualization and analysis tools that help reveal higher-level patterns in the underlying data. The current iterations of TCRD and Pharos enable users to perform enrichment calculations based on subsets of targets, diseases, or ligands and to create interactive heat maps and UpSet charts of many types of annotations. Using several examples, we show how to address disease biology and drug discovery questions through enrichment calculations and UpSet charts.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Edwards A.M., Isserlin R., Bader G.D., Frye S.V., Willson T.M., Yu F.H.. Too many roads not taken. Nature. 2011; 470:163–165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

