GliaMorph: a modular image analysis toolkit to quantify Müller glial cell morphology
- PMID: 36625162
- PMCID: PMC10110500
- DOI: 10.1242/dev.201008
GliaMorph: a modular image analysis toolkit to quantify Müller glial cell morphology
Abstract
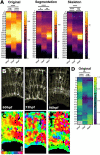
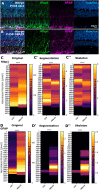
Cell morphology is crucial for all cell functions. This is particularly true for glial cells as they rely on complex shape to contact and support neurons. However, methods to quantify complex glial cell shape accurately and reproducibly are lacking. To address this, we developed the image analysis pipeline 'GliaMorph'. GliaMorph is a modular analysis toolkit developed to perform (1) image pre-processing, (2) semi-automatic region-of-interest selection, (3) apicobasal texture analysis, (4) glia segmentation, and (5) cell feature quantification. Müller glia (MG) have a stereotypic shape linked to their maturation and physiological status. Here, we characterized MG on three levels: (1) global image-level, (2) apicobasal texture, and (3) regional apicobasal vertical-to-horizontal alignment. Using GliaMorph, we quantified MG development on a global and single-cell level, showing increased feature elaboration and subcellular morphological rearrangement in the zebrafish retina. As proof of principle, we analysed expression changes in a mouse glaucoma model, identifying subcellular protein localization changes in MG. Together, these data demonstrate that GliaMorph enables an in-depth understanding of MG morphology in the developing and diseased retina.
Keywords: Development; Glia morphology; Mouse; Müller glia; Retina; Zebrafish.
© 2023. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures







References
-
- Ali, F. (2007). Image restoration using regularized inverse filtering and adaptive threshold wavelet denoising. Al-Khwarizmi Engineering Journal 3, 48-62.
-
- Beck, A. and Teboulle, M. (2009). A fast iterative shrinkage-thresholding algorithm for linear inverse problems. SIAM J. Imaging Sci. 2, 183-202. 10.1137/080716542 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

