Genomic Epidemiology and Multilevel Genome Typing of Australian Salmonella enterica Serovar Enteritidis
- PMID: 36625638
- PMCID: PMC9927265
- DOI: 10.1128/spectrum.03014-22
Genomic Epidemiology and Multilevel Genome Typing of Australian Salmonella enterica Serovar Enteritidis
Abstract
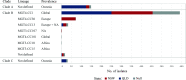
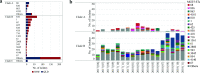
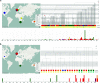
Salmonella enterica serovar Enteritidis is one of the leading causes of salmonellosis in Australia. In this study, a total of 568 S. Enteritidis isolates from two Australian states across two consecutive years were analyzed and compared to international strains, using the S. Enteritidis multilevel genome typing (MGT) database, which contained 40,390 publicly available genomes from 99 countries. The Australian S. Enteritidis isolates were divided into three phylogenetic clades (A, B, and C). Clades A and C represented 16.4% and 3.5% of the total isolates, respectively, and were of local origin. Clade B accounted for 80.1% of the isolates which belonged to seven previously defined lineages but was dominated by the global epidemic lineage. At the MGT5 level, three out of five top sequence types (STs) in Australia were also top STs in Asia, suggesting that a fair proportion of Australian S. Enteritidis cases may be epidemiologically linked with Asian strains. In 2018, a large egg-associated local outbreak was caused by a recently defined clade B lineage prevalent in Europe and was closely related, but not directly linked, to three European isolates. Additionally, over half (54.8%) of predicted multidrug resistance (MDR) isolates belonged to 10 MDR-associated MGT-STs, which were also frequent in Asian S. Enteritidis . Overall, this study investigated the genomic epidemiology of S. Enteritidis in Australia, including the first large local outbreak, using MGT. The open MGT platform enables a standardized and sharable nomenclature that can be effectively applied to public health for unified surveillance of S. Enteritidis nationally and globally. IMPORTANCE Salmonella enterica serovar Enteritidis is a leading cause of foodborne infections. We previously developed a genomic typing database (MGTdb) for S. Enteritidis to facilitate global surveillance of this pathogen. In this study, we examined the genomic features of Australian S. Enteritidis using the MGTdb and found that Australian S. Enteritidis is mainly epidemiologically linked with Asian strains (especially strains carrying antimicrobial resistance genes), followed by European strains. The first large-scale egg-associated local outbreak in Australia was caused by a recently defined lineage prevalent in Europe, and three European isolates in the MGTdb were closely related but not directly linked to this outbreak. In summary, the S. Enteritidis MGTdb open platform is shown to be a potentially powerful tool for national and global public health surveillance of this pathogen.
Keywords: Salmonella Enteritidis; foodborne outbreak; genomic epidemiology; genomic typing database; multilevel genome typing; standardized genomic typing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, Krauland MG, Hale JL, Harbottle H, Uesbeck A, Dougan G, Harrison LH, Brisse S, S. Enterica MLST Study Group . 2012. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog 8:e1002776. doi:10.1371/journal.ppat.1002776. - DOI - PMC - PubMed
-
- Feasey NA, Hadfield J, Keddy KH, Dallman TJ, Jacobs J, Deng X, Wigley P, Barquist L, Langridge GC, Feltwell T, Harris SR, Mather AE, Fookes M, Aslett M, Msefula C, Kariuki S, Maclennan CA, Onsare RS, Weill FX, Le Hello S, Smith AM, McClelland M, Desai P, Parry CM, Cheesbrough J, French N, Campos J, Chabalgoity JA, Betancor L, Hopkins KL, Nair S, Humphrey TJ, Lunguya O, Cogan TA, Tapia MD, Sow SO, Tennant SM, Bornstein K, Levine MM, Lacharme-Lora L, Everett DB, Kingsley RA, Parkhill J, Heyderman RS, Dougan G, Gordon MA, Thomson NR. 2016. Distinct Salmonella Enteritidis lineages associated with enterocolitis in high-income settings and invasive disease in low-income settings. Nat Genet 48:1211–1217. doi:10.1038/ng.3644. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

