The Substitutions L50F, E166A, and L167F in SARS-CoV-2 3CLpro Are Selected by a Protease Inhibitor In Vitro and Confer Resistance To Nirmatrelvir
- PMID: 36625640
- PMCID: PMC9973015
- DOI: 10.1128/mbio.02815-22
The Substitutions L50F, E166A, and L167F in SARS-CoV-2 3CLpro Are Selected by a Protease Inhibitor In Vitro and Confer Resistance To Nirmatrelvir
Abstract
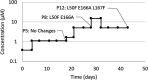
The SARS-CoV-2 main protease (3CLpro) has an indispensable role in the viral life cycle and is a therapeutic target for the treatment of COVID-19. The potential of 3CLpro-inhibitors to select for drug-resistant variants needs to be established. Therefore, SARS-CoV-2 was passaged in vitro in the presence of increasing concentrations of ALG-097161, a probe compound designed in the context of a 3CLpro drug discovery program. We identified a combination of amino acid substitutions in 3CLpro (L50F E166A L167F) that is associated with a >20× increase in 50% effective concentration (EC50) values for ALG-097161, nirmatrelvir (PF-07321332), PF-00835231, and ensitrelvir. While two of the single substitutions (E166A and L167F) provide low-level resistance to the inhibitors in a biochemical assay, the triple mutant results in the highest levels of resistance (6× to 72×). All substitutions are associated with a significant loss of enzymatic 3CLpro activity, suggesting a reduction in viral fitness. Structural biology analysis indicates that the different substitutions reduce the number of inhibitor/enzyme interactions while the binding of the substrate is maintained. These observations will be important for the interpretation of resistance development to 3CLpro inhibitors in the clinical setting. IMPORTANCE Paxlovid is the first oral antiviral approved for treatment of SARS-CoV-2 infection. Antiviral treatments are often associated with the development of drug-resistant viruses. In order to guide the use of novel antivirals, it is essential to understand the risk of resistance development and to characterize the associated changes in the viral genes and proteins. In this work, we describe for the first time a pathway that allows SARS-CoV-2 to develop resistance against Paxlovid in vitro. The characteristics of in vitro antiviral resistance development may be predictive for the clinical situation. Therefore, our work will be important for the management of COVID-19 with Paxlovid and next-generation SARS-CoV-2 3CLpro inhibitors.
Keywords: antiviral agents; coronavirus; drug resistance mechanisms; protease inhibitors.
Conflict of interest statement
The authors declare a conflict of interest. The authors declare the following financial interests/personal relationships which may be considered as potential competing interests. Koen Vandyck and Pierre Raboisson are employees of Aligos Belgium BV. Cheng Liu, Antitsa Stoycheva, Sarah K Stevens, Chloe De Vita, Andreas Jekle, Lawrence M Blatt, Leonid Beigelman, Julian A Symons and Jerome Deval are employees of Aligos Therapeutics, Inc. A patent application on ALG-097161 is pending.
Figures
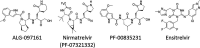
References
-
- Owen DR, Allerton CMN, Anderson AS, Aschenbrenner L, Avery M, Berritt S, Boras B, Cardin RD, Carlo A, Coffman KJ, Dantonio A, Di L, Eng H, Ferre R, Gajiwala KS, Gibson SA, Greasley SE, Hurst BL, Kadar EP, Kalgutkar AS, Lee JC, Lee J, Liu W, Mason SW, Noell S, Novak JJ, Obach RS, Ogilvie K, Patel NC, Pettersson M, Rai DK, Reese MR, Sammons MF, Sathish JG, Singh RSP, Steppan CM, Stewart AE, Tuttle JB, Updyke L, Verhoest PR, Wei L, Yang Q, Zhu Y. 2021. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science 374:1586–1593. doi:10.1126/science.abl4784. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

