Epitranscriptomic N6-Methyladenosine Profile of SARS-CoV-2-Infected Human Lung Epithelial Cells
- PMID: 36625663
- PMCID: PMC9927293
- DOI: 10.1128/spectrum.03943-22
Epitranscriptomic N6-Methyladenosine Profile of SARS-CoV-2-Infected Human Lung Epithelial Cells
Abstract
N6-methyladenosine (m6A) is a dynamic posttranscriptional RNA modification that plays an important role in determining transcript fate. The functional consequence of m6A deposition is dictated by a group of host proteins that specifically recognize and bind the m6A modification, leading to changes in RNA stability, transport, splicing, or translation. The cellular m6A methylome undergoes changes during certain pathogenic conditions such as viral infections. However, how m6A modification of host cell transcripts and noncoding RNAs change during severe acute respiratory syndrome coronavirus (SARS-CoV-2) infection has not been reported. Here, we define the epitranscriptomic m6A profile of SARS-CoV-2-infected human lung epithelial cells compared to uninfected controls. We identified mRNA and long and small noncoding RNA species that are differentially m6A modified in response to SARS-CoV-2 infection. The most significantly differentially methylated transcript was the precursor of microRNA-4486 (miRNA-4486), which showed significant increases in abundance and percentage of methylated transcripts in infected cells. Pathway analyses revealed that differentially methylated transcripts were significantly associated with several cancer-related pathways, protein processing in the endoplasmic reticulum, cell death, and proliferation. Upstream regulators predicted to be associated with the proteins encoded by differentially methylated mRNAs include several proteins involved in the type-I interferon response, inflammation, and cytokine signaling. IMPORTANCE Posttranscriptional modification of viral and cellular RNA by N6-methyladenosine (m6A) plays an important role in regulating the replication of many viruses and the cellular immune response to infection. We therefore sought to define the epitranscriptomic m6A profile of human lung epithelial cells infected with SARS-CoV-2. Our analyses demonstrate the differential methylation of both coding and noncoding cellular RNAs in SARS-CoV-2-infected cells compared to uninfected controls. Pathway analyses revealed that several of these RNAs may be involved in the cellular response to infection, such as type-I interferon. Our study implicates m6A modification of infected-cell RNA as a mechanism of posttranscriptional gene regulation during SARS-CoV-2 infection.
Keywords: N6-methyladenosine; SARS-CoV-2; epitranscriptomics; infection; lung epithelial cells; microarray.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

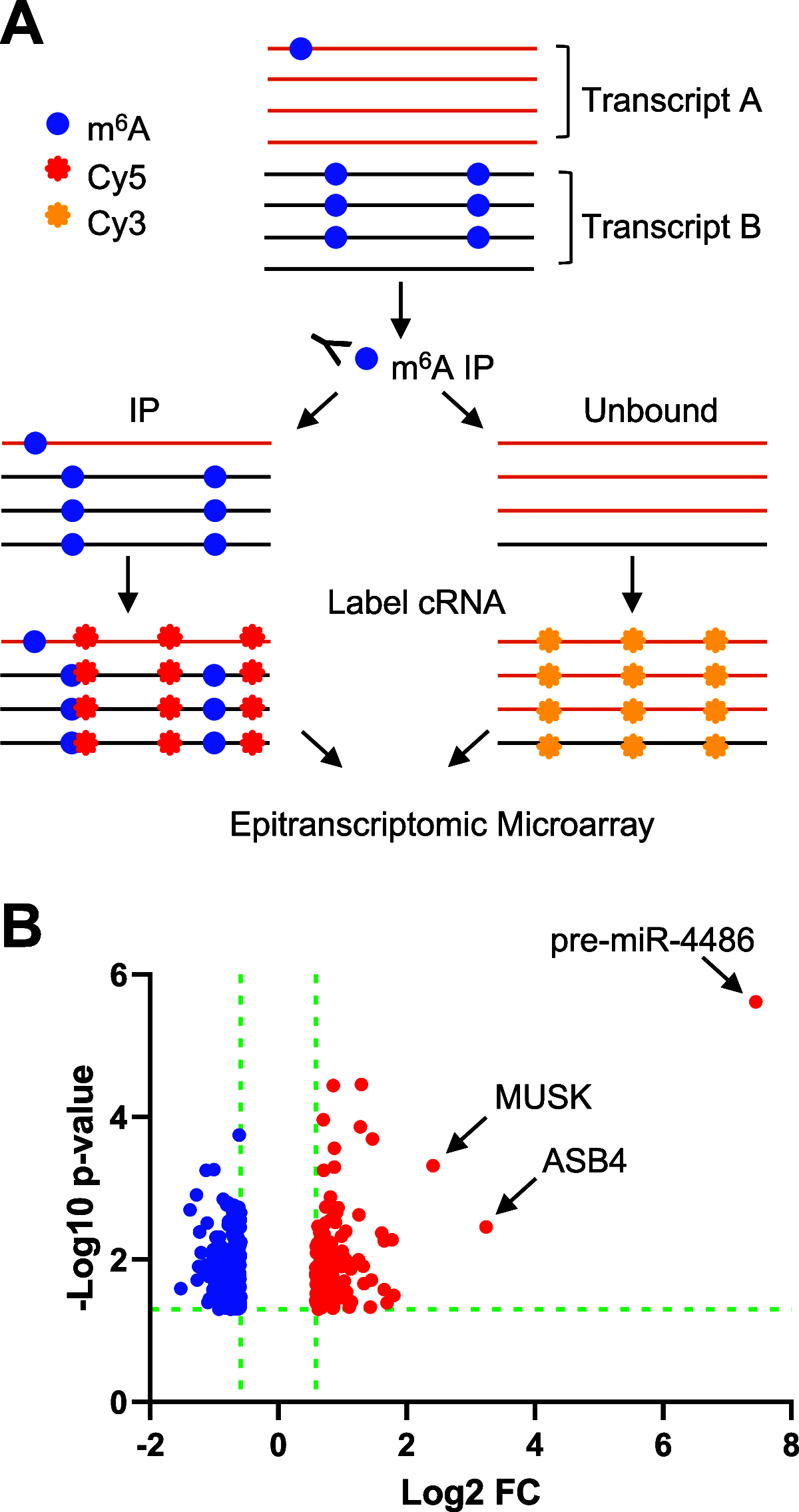
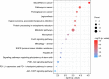

References
-
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, Lu Z, Bosmans RP, Dai Q, Hao YJ, Yang X, Zhao WM, Tong WM, Wang XJ, Bogdan F, Furu K, Fu Y, Jia G, Zhao X, Liu J, Krokan HE, Klungland A, Yang YG, He C. 2013. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49:18–29. doi:10.1016/j.molcel.2012.10.015. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

