Multiscale molecular dynamics simulations predict arachidonic acid binding sites in human ASIC1a and ASIC3 transmembrane domains
- PMID: 36625864
- PMCID: PMC9836442
- DOI: 10.1085/jgp.202213259
Multiscale molecular dynamics simulations predict arachidonic acid binding sites in human ASIC1a and ASIC3 transmembrane domains
Abstract
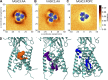
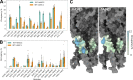
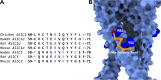
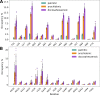
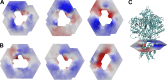
Acid-sensing ion channels (ASICs) play important roles in inflammatory pathways by conducting ions across the neuronal membrane in response to proton binding under acidic conditions. Recent studies have shown that ASICs can be modulated by arachidonic acid (AA), and, in the case of the ASIC3 subtype, even activated by AA at physiological pH. However, the mechanism by which these fatty acids act on the channel is still unknown. Here, we have used multiscale molecular dynamics simulations to predict a putative, general binding region of AA to models of the human ASIC protein. We have identified, in agreement with recent studies, residues in the outer leaflet transmembrane region which interact with AA. In addition, despite their similar modulation, we observe subtle differences in the AA interaction pattern between human ASIC1a and human ASIC3, which can be reversed by mutating three key residues at the outer leaflet portion of TM1. We further probed interactions with these residues in hASIC3 using atomistic simulations and identified possible AA coordinating interactions; salt bridge interactions of AA with R65hASIC3 and R68hASIC3 and AA tail interactions with the Y58hASIC3 aromatic ring. We have shown that longer fatty acid tails with more double bonds have increased relative occupancy in this region of the channel, a finding supported by recent functional studies. We further proposed that the modulatory effect of AA on ASIC does not result from changes in local membrane curvature. Rather, we speculate that it may occur through structural changes to the ion channel upon AA binding.
© 2023 Ananchenko and Musgaard.
Figures






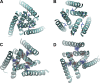
Comment in
-
A loosely coordinated interaction site for arachidonic acid on ASICs.J Gen Physiol. 2023 Mar 6;155(3):e202213307. doi: 10.1085/jgp.202213307. Epub 2023 Feb 1. J Gen Physiol. 2023. PMID: 36723670 Free PMC article.
Similar articles
-
Identification of the modulatory Ca2+-binding sites of acid-sensing ion channel 1a.Open Biol. 2024 Jun;14(6):240028. doi: 10.1098/rsob.240028. Epub 2024 Jun 19. Open Biol. 2024. PMID: 38896086 Free PMC article.
-
Heteroarylguanidines as Allosteric Modulators of ASIC1a and ASIC3 Channels.ACS Chem Neurosci. 2018 Jun 20;9(6):1357-1365. doi: 10.1021/acschemneuro.7b00529. Epub 2018 Mar 30. ACS Chem Neurosci. 2018. PMID: 29566331
-
Pharmacological modulation of Acid-Sensing Ion Channels 1a and 3 by amiloride and 2-guanidine-4-methylquinazoline (GMQ).Neuropharmacology. 2017 Oct;125:429-440. doi: 10.1016/j.neuropharm.2017.08.004. Epub 2017 Aug 9. Neuropharmacology. 2017. PMID: 28802647
-
Acid-Sensing Ion Channels: Focus on Physiological and Some Pathological Roles in the Brain.Curr Neuropharmacol. 2021;19(9):1570-1589. doi: 10.2174/1570159X19666210125151824. Curr Neuropharmacol. 2021. PMID: 33550975 Free PMC article. Review.
-
Peptides inhibitors of acid-sensing ion channels.Toxicon. 2007 Feb;49(2):271-84. doi: 10.1016/j.toxicon.2006.09.026. Epub 2006 Oct 4. Toxicon. 2007. PMID: 17113616 Review.
Cited by
-
State-dependent binding of cholesterol and an anionic lipid to the muscle-type Torpedo nicotinic acetylcholine receptor.Commun Biol. 2024 Apr 10;7(1):437. doi: 10.1038/s42003-024-06106-8. Commun Biol. 2024. PMID: 38600247 Free PMC article.
-
Molecular Insights into Single Chain Lipid Modulation of Acid-Sensing Ion Channel 3.bioRxiv [Preprint]. 2024 Aug 30:2024.08.29.610156. doi: 10.1101/2024.08.29.610156. bioRxiv. 2024. Update in: J Phys Chem B. 2024 Dec 26;128(51):12685-12697. doi: 10.1021/acs.jpcb.4c04289. PMID: 39257759 Free PMC article. Updated. Preprint.
-
A loosely coordinated interaction site for arachidonic acid on ASICs.J Gen Physiol. 2023 Mar 6;155(3):e202213307. doi: 10.1085/jgp.202213307. Epub 2023 Feb 1. J Gen Physiol. 2023. PMID: 36723670 Free PMC article.
References
-
- Abraham, M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., and Lindahl E.. 2015. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1–2:19–25. 10.1016/j.softx.2015.06.001 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

