microRNA-33 deficiency in macrophages enhances autophagy, improves mitochondrial homeostasis, and protects against lung fibrosis
- PMID: 36626225
- PMCID: PMC9977502
- DOI: 10.1172/jci.insight.158100
microRNA-33 deficiency in macrophages enhances autophagy, improves mitochondrial homeostasis, and protects against lung fibrosis
Abstract
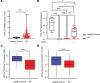
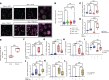
Idiopathic pulmonary fibrosis (IPF) is a progressive and ultimately fatal disease. Recent findings have shown a marked metabolic reprogramming associated with changes in mitochondrial homeostasis and autophagy during pulmonary fibrosis. The microRNA-33 (miR-33) family of microRNAs (miRNAs) encoded within the introns of sterol regulatory element binding protein (SREBP) genes are master regulators of sterol and fatty acid (FA) metabolism. miR-33 controls macrophage immunometabolic response and enhances mitochondrial biogenesis, FA oxidation, and cholesterol efflux. Here, we show that miR-33 levels are increased in bronchoalveolar lavage (BAL) cells isolated from patients with IPF compared with healthy controls. We demonstrate that specific genetic ablation of miR-33 in macrophages protects against bleomycin-induced pulmonary fibrosis. The absence of miR-33 in macrophages improves mitochondrial homeostasis and increases autophagy while decreasing inflammatory response after bleomycin injury. Notably, pharmacological inhibition of miR-33 in macrophages via administration of anti-miR-33 peptide nucleic acids (PNA-33) attenuates fibrosis in different in vivo and ex vivo mice and human models of pulmonary fibrosis. These studies elucidate a major role of miR-33 in macrophages in the regulation of pulmonary fibrosis and uncover a potentially novel therapeutic approach to treat this disease.
Keywords: Autophagy; Fatty acid oxidation; Fibrosis; Metabolism; Pulmonology.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

