SHP2 deneddylation mediates tumor immunosuppression in colon cancer via the CD47/SIRPα axis
- PMID: 36626230
- PMCID: PMC9927946
- DOI: 10.1172/JCI162870
SHP2 deneddylation mediates tumor immunosuppression in colon cancer via the CD47/SIRPα axis
Abstract
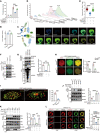
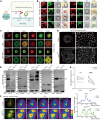
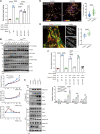
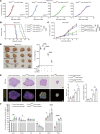
SIPRα on macrophages binds with CD47 to resist proengulfment signals, but how the downstream signal of SIPRα controls tumor-infiltrating macrophages (TIMs) is still poorly clarified. Here, we report that the CD47/signal regulatory protein α (SIRPα) axis requires the deneddylation of tyrosine phosphatase SHP2. Mechanistically, Src homology region 2-containing protein tyrosine phosphatase 2 (SHP2) was constitutively neddylated on K358 and K364 sites; thus, its autoinhibited conformation was maintained. In response to CD47-liganded SIRPα, SHP2 was deneddylated by sentrin-specific protease 8 (SENP8), which led to the dephosphorylation of relevant substrates at the phagocytic cup and subsequent inhibition of macrophage phagocytosis. Furthermore, neddylation inactivated myeloid-SHP2 and greatly boosted the efficacy of colorectal cancer (CRC) immunotherapy. Importantly, we observed that supplementation with SHP2 allosteric inhibitors sensitized immune treatment-resistant CRC to immunotherapy. Our results emphasize that the CRC subtype that is unresponsive to immunotherapy relies on SIRPαhiSHP2hiNEDD8lo TIMs and highlight the need to further explore the strategy of SHP2 targeting in CRC therapy.
Keywords: Cell Biology; Immunotherapy; Macrophages; Signal transduction.
Figures







Comment in
- Waking immune-resistant tumors with neddlyation
References
-
- Wei SC, et al. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018;8(9):1069–1086. doi: 10.1158/2159-8290.CD-18-0367. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

