Population-specific positive selection on low CR1 expression in malaria-endemic regions
- PMID: 36626386
- PMCID: PMC9831336
- DOI: 10.1371/journal.pone.0280282
Population-specific positive selection on low CR1 expression in malaria-endemic regions
Abstract
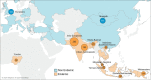
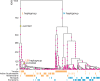
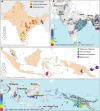
Complement Receptor Type 1 (CR1) is a malaria-associated gene that encodes a transmembrane receptor of erythrocytes and is crucial for malaria parasite invasion. The expression of CR1 contributes to the rosetting of erythrocytes in the brain bloodstream, causing cerebral malaria, the most severe form of the disease. Here, we study the history of adaptation against malaria by analyzing selection signals in the CR1 gene. We used whole-genome sequencing datasets of 907 healthy individuals from malaria-endemic and non-endemic populations. We detected robust positive selection in populations from the hyperendemic regions of East India and Papua New Guinea. Importantly, we identified a new adaptive variant, rs12034598, which is associated with a slower rate of erythrocyte sedimentation and is linked with a variant associated with low levels of CR1 expression. The combination of the variants likely drives natural selection. In addition, we identified a variant rs3886100 under positive selection in West Africans, which is also related to a low level of CR1 expression in the brain. Our study shows the fine-resolution history of positive selection in the CR1 gene and suggests a population-specific history of CR1 adaptation to malaria. Notably, our novel approach using population genomic analyses allows the identification of protective variants that reduce the risk of malaria infection without the need for patient samples or malaria individual medical records. Our findings contribute to understanding of human adaptation against cerebral malaria.
Copyright: © 2023 Lorenzini et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Santoro F, Bernal J, Capron A. Complement activation by parasites. A review. Acta Trop. 1979;36(1):5–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

