CITED2 is a conserved regulator of the uterine-placental interface
- PMID: 36626551
- PMCID: PMC9934066
- DOI: 10.1073/pnas.2213622120
CITED2 is a conserved regulator of the uterine-placental interface
Abstract
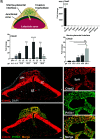
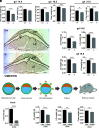
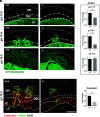
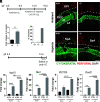
Establishment of the hemochorial uterine-placental interface requires exodus of trophoblast cells from the placenta and their transformative actions on the uterus, which represent processes critical for a successful pregnancy, but are poorly understood. We examined the involvement of CBP/p300-interacting transactivator with glutamic acid/aspartic acid-rich carboxyl-terminal domain 2 (CITED2) in rat and human trophoblast cell development. The rat and human exhibit deep hemochorial placentation. CITED2 was distinctively expressed in the junctional zone (JZ) and invasive trophoblast cells of the rat. Homozygous Cited2 gene deletion resulted in placental and fetal growth restriction. Small Cited2 null placentas were characterized by disruptions in the JZ, delays in intrauterine trophoblast cell invasion, and compromised plasticity. In the human placentation site, CITED2 was uniquely expressed in the extravillous trophoblast (EVT) cell column and importantly contributed to the development of the EVT cell lineage. We conclude that CITED2 is a conserved regulator of deep hemochorial placentation.
Keywords: CITED2; placenta; pregnancy; stem cells; trophoblast.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Maltepe E., Fisher S. J., Placenta: The forgotten organ. Annu. Rev. Cell Dev. Biol. 31, 523–552 (2015). - PubMed
-
- Pijnenborg R., Robertson W. B., Brosens I., Dixon G., Trophoblast invasion and the establishment of haemochorial placentation in man and laboratory animals. Placenta 2, 71–91 (1981). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

