IRF8 and its related molecules as potential diagnostic biomarkers or therapeutic candidates and immune cell infiltration characteristics in steroid-induced osteonecrosis of the femoral head
- PMID: 36627660
- PMCID: PMC9832881
- DOI: 10.1186/s13018-022-03381-1
IRF8 and its related molecules as potential diagnostic biomarkers or therapeutic candidates and immune cell infiltration characteristics in steroid-induced osteonecrosis of the femoral head
Abstract
Purpose: Steroid-induced osteonecrosis of the femoral head (SONFH) was a refractory orthopedic hip joint disease in the young and middle-aged people, but the pathogenesis of SONFH remained unclear. We aimed to identify the potential genes and screen potential therapeutic compounds for SONFH.
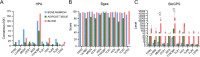
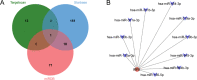
Methods: The microarray was obtained for blood tissue from the GEO database, and then it identifies differentially expressed genes (DEGs). The DEGs were analyzed to obtain the differences in immune cell infiltration. The gene functional enrichment analysis of SONFH was analyzed. The PPI of DEGs was identified through the STRING database, and the cluster modules and hub genes were ascertained using MCODE and CytoHubba, and the ROC curve of hub genes was analyzed, and the tissues distribution of hub genes was understood by the HPA, Bgee and BioGPS databases. The hub genes and target miRNAs and corresponding upstream lncRNAs were predicted by TargetScan, miRDB and ENCORI database. Subsequently, we used CMap, DGIdb and L1000FWD databases to identify several potential therapeutic molecular compounds for SONFH. Finally, the AutoDockTools Vina, PyMOL and Discovery Studio were employed for molecular docking analyses between compounds and hub genes.
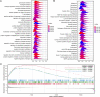
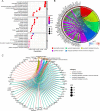
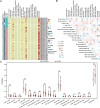
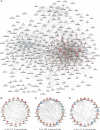
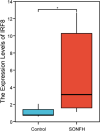
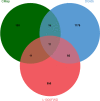
Results: The microarray dataset GSE123568 was obtained related to SONFH. There were 372 DEGs including 197 upregulated genes and 175 downregulated genes by adjusted P value < 0.01 and |log2FC|> 1. Several significant GSEA enrichment analysis and biological processes and KEGG pathway associated with SONFH were identified, which were significantly related to cytoskeleton organization, nucleobase-containing compound catabolic process, NOD-like receptor signaling pathway, MAPK signaling pathway, FoxO signaling pathway, neutrophil-mediated immunity, neutrophil degranulation and neutrophil activation involved in immune response. Activated T cells CD4 memory, B cells naïve, B cells memory, T cells CD8 and T cells gamma delta might be involved in the occurrence and development of SONFH. Three cluster modules were identified in the PPI network, and eleven hub genes including FPR2, LILRB2, MNDA, CCR1, IRF8, TYROBP, TLR1, HCK, TLR8, TLR2 and CCR2 were identified by Cytohubba, which were differed in bone marrow, adipose tissue and blood, and which had good diagnostic performance in SONFH. We identified IRF8 and 10 target miRNAs that was utilized including Targetsan, miRDB and ENCORI databases and 8 corresponding upstream lncRNAs that was revealed by ENCORI database. IRF8 was detected with consistent expression by qRT-PCR. Based on the CMap, DGIdb and L1000FWD databases, the 11 small molecular compounds that were most strongly therapeutic correlated with SONFH were estradiol, genistein, domperidone, lovastatin, myricetin, fenbufen, rosiglitazone, sirolimus, phenformin, vorinostat and vinblastine. All of 11 small molecules had good binding affinity with the IRF8 in molecular docking.
Conclusion: The occurrence of SONFH was associated with a "multi-target" and "multi-pathway" pattern, especially related to immunity, and IRF8 and its noncoding RNA were closely related to the development of SONFH. The CMap, DGIdb and L1000FWD databases could be effectively used in a systematic manner to predict potential drugs for the prevention and treatment of SONFH. However, additional clinical and experimental research is warranted.
Keywords: Competing endogenous RNA; Connectivity map database; Drug gene interaction database; Immune cell infiltration; L1000 fireworks display database; Molecular docking; Noncoding RNA; Steroid-induced osteonecrosis of the femoral head.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest regarding this work.
Figures






References
MeSH terms
Substances
Grants and funding
- No. 81774333/National Natural Science Foundation of China
- No. 82074453/National Natural Science Foundation of China
- No. ZR2021QH004/National Natural Science Foundation of Shandong Province
- No. 2019WS577/Development Plan of Shandong Medical and Health Technology
- No. 2020Q009/Development Plan of Shandong Traditional Chinese Medicine Science and Technology
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

