OKseqHMM: a genome-wide replication fork directionality analysis toolkit
- PMID: 36629249
- PMCID: PMC9976876
- DOI: 10.1093/nar/gkac1239
OKseqHMM: a genome-wide replication fork directionality analysis toolkit
Abstract
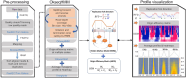
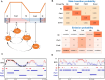
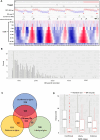
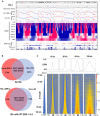
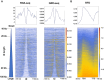
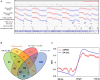
During each cell division, tens of thousands of DNA replication origins are co-ordinately activated to ensure the complete duplication of the human genome. However, replication fork progression can be challenged by many factors, including co-directional and head-on transcription-replication conflicts (TRC). Head-on TRCs are more dangerous for genome integrity. To study the direction of replication fork movement and TRCs, we developed a bioinformatics toolkit called OKseqHMM (https://github.com/CL-CHEN-Lab/OK-Seq, https://doi.org/10.5281/zenodo.7428883). Then, we used OKseqHMM to analyse a large number of datasets obtained by Okazaki fragment sequencing to directly measure the genome-wide replication fork directionality (RFD) and to accurately predict replication initiation and termination at a fine resolution in organisms including yeast, mouse and human. We also successfully applied our analysis to other genome-wide sequencing techniques that also contain RFD information (e.g. eSPAN, TrAEL-seq). Our toolkit can be used to predict replication initiation and fork progression direction genome-wide in a wide range of cell models and growth conditions. Comparing the replication and transcription directions allows identifying loci at risk of TRCs, particularly head-on TRCs, and investigating their role in genome instability by checking DNA damage data, which is of prime importance for human health.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Gnan S., Liu Y., Spagnuolo M., Chen C.-L. The impact of transcription-mediated replication stress on genome instability and human disease. Genome Instab. Dis. 2020; 1:207–234.
-
- Chen C.-L., Duquenne L., Audit B., Guilbaud G., Rappailles A., Baker A., Huvet M., D’Aubenton-Carafa Y., Hyrien O., Arneodo A. et al. . Replication-associated mutational asymmetry in the human genome. Mol. Biol. Evol. 2011; 28:2327–2337. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

