TIMP-1 is a novel ligand of Amyloid Precursor Protein and triggers a proinflammatory phenotype in human monocytes
- PMID: 36629908
- PMCID: PMC9837626
- DOI: 10.1083/jcb.202206095
TIMP-1 is a novel ligand of Amyloid Precursor Protein and triggers a proinflammatory phenotype in human monocytes
Abstract
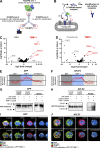
The emerging cytokine tissue inhibitor of metalloproteinases-1 (TIMP-1) correlates with the progression of inflammatory diseases, including cancer. However, the effects of TIMP-1 on immune cell activation and underlying molecular mechanisms are largely unknown. Unbiased ligand-receptor-capture-screening revealed TIMP-1-interaction with Amyloid Precursor Protein (APP) family members, namely APP and Amyloid Precursor-like Protein-2 (APLP2), which was confirmed by pull-down assays and confocal microscopy. We found that TIMP-1 triggered glucose uptake and proinflammatory cytokine expression in human monocytes. In cancer patients, TIMP-1 expression positively correlated with proinflammatory cytokine expression and processes associated with monocyte activation. In pancreatic cancer, TIMP-1 plasma levels correlated with the monocyte activation marker sCD163, and the combined use of both clinically accessible plasma proteins served as a powerful prognostic indicator. Mechanistically, TIMP-1 triggered monocyte activation by its C-terminal domain and via APP as demonstrated by in vitro interference, in silico docking, and the employment of recombinant TIMP-1 variants. Identification of TIMP-1 as a trigger of monocyte activation opens new therapeutic perspectives for inflammatory diseases.
© 2023 Eckfeld et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures






References
-
- Bruchez, A., Sha K., Johnson J., Chen L., Stefani C., McConnell H., Gaucherand L., Prins R., Matreyek K.A., Hume A.J., et al. 2020. MHC class II transactivator CIITA induces cell resistance to Ebola virus and SARS-like coronaviruses. Science. 370:241–247. 10.1126/science.abb3753 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

