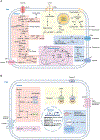
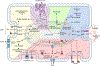
Figure 2.. Macronutrient uptake and metabolism control of TAM responses
A) Macronutrients including glucose, lipids and amino acids are taken up by tumor-associated macrophages (TAMs) in the tumor microenvironment (TME) and are catabolized or converted to biosynthetic intermediates or signaling metabolites to regulate TAM responses. Glucose acquired through glucose transporter 1 (GLUT1) undergoes glycolysis to produce adenosine triphosphate (ATP) and generates metabolic intermediates to support several biosynthetic pathways. Pyruvate is the end-product of glycolysis, and is mainly reduced to lactate in the cytosol, rather than enter the mitochondrion to complete the tricarboxylic acid (TCA) cycle as a likely consequence of low oxygen level in the TME. Instead, itaconate can be converted from the TCA intermediate cis-aconitate with tumor-promoting functions. Long-chain fatty acids (LCFAs) are acquired via CD36-mediated lipid uptake and contribute to lipogenesis and lipid droplet formation in TAMs, while prostaglandin E2 (PGE2) and leukotrienes are converted from phospholipids, and act as bioactive signaling lipids. Amino acids arginine and tryptophan acquired from plasma membrane transporters can also be converted to bioactive metabolites to regulate tumor progression. B) Signaling functions of metabolites in TAMs. Metabolites of the glycolytic pathway can promote activation of transcription factors including signal transducer and activator of transcription 6 (STAT6), nuclear factor kappa B (NF-κB), and hypoxia-inducible factor 1-alpha (HIF-1α). In addition, lactate can be sensed by the G protein-coupled receptors (GPCRs) GPR132 and OLFR78, while the TCA cycle metabolite itaconate suppresses the generation of reactive oxygen species (ROS). Lipid-sensing peroxisome proliferator-activated receptor-γ (PPAR-γ) is subject to caspase-1-mediated cleavage to prevent fatty acid oxidation, while lipid-derived metabolites including vitamin D, PGE2, and leukotrienes can bind to vitamin D receptor (VDR) and GPCR family members to induce cellular signaling. Moreover, cytosolic amino acids such as leucine promotes activation of the metabolic regulator mammalian target of rapamycin complex 1 (mTORC1), while the tryptophan metabolites kynurenine, kynurenic acid, and indole metabolites are sensed by aryl hydrocarbon receptor (AhR) and GPCR family member to regulate TAM responses. 1,3BPG, 1,3-bisphosphoglycerate; 3-PG, 3-phosphoglycerate; 5-LO, 5-lipoxygenase; α-KG, alpha-ketoglutarate; ADP, adenosine diphosphate; Arg1, arginase 1; CAT, cationic amino acid transporter; CCL, chemokine (C-C motif) ligand; COX, cyclooxygenase; CXCL, chemokine (C-X-C motif) ligand; DAGs, diacylglycerols; DGAT, diglyceride acyltransferase; DHAP, dihydroxyacetone phosphate; ETC, electron transport chain; F1,6BP, fructose-1,6-bisphosphate; F2,6BP, fructose-2,6-bisphosphate; F6P, fructose 6-phosphate; FABPs, fatty acid-binding proteins; G3P, glycerol 3-phosphate; G6P, glucose 6-phosphate; GA3P, glyceraldehyde 3-phosphate; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IL, interleukin; IL4I1, interleukin-4 induced 1; IRG1, immune-responsive gene 1; LDH-A, lactate dehydrogenase-A; MAGs, monoacylglycerols; MCT, monocarboxylate transporter; MGLL, monoacylglycerol lipase; MMPs, matrix metalloproteinase; NAD, nicotinamide adenine dinucleotide; NADP, nicotinamide adenine dinucleotide phosphate; OAA, oxaloacetate; PC, pyruvate carboxylase; PDH, pyruvate dehydrogenase; PDK1, pyruvate dehydrogenase kinase 1; PD-L1, programmed death-ligand 1; PEP, phosphoenolpyruvate; PFK-1, 6-phosphofructokinase-1; PFKFB3, 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3; PKM2, pyruvate kinase M2; PSAT, phosphoserine aminotransferase; Ribose-5P, ribose 5-phosphate; RXR, retinoid-X receptor; TAGs, triacylglycerols; TFs, transcription factors; TGF-β, transforming growth factor-beta; UDP-GlcNAc, uridine diphosphate-N-acetylglucosamine; VEGF-α, vascular endothelial growth factor-alpha.