GATA4 controls regionalization of tissue immunity and commensal-driven immunopathology
- PMID: 36630917
- PMCID: PMC10262782
- DOI: 10.1016/j.immuni.2022.12.009
GATA4 controls regionalization of tissue immunity and commensal-driven immunopathology
Abstract
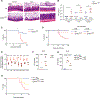
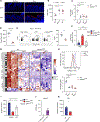
There is growing recognition that regionalization of bacterial colonization and immunity along the intestinal tract has an important role in health and disease. Yet, the mechanisms underlying intestinal regionalization and its dysregulation in disease are not well understood. This study found that regional epithelial expression of the transcription factor GATA4 controls bacterial colonization and inflammatory tissue immunity in the proximal small intestine by regulating retinol metabolism and luminal IgA. Furthermore, in mice without jejunal GATA4 expression, the commensal segmented filamentous bacteria promoted pathogenic inflammatory immune responses that disrupted barrier function and increased mortality upon Citrobacter rodentium infection. In celiac disease patients, low GATA4 expression was associated with metabolic alterations, mucosal Actinobacillus, and increased IL-17 immunity. Taken together, these results reveal broad impacts of GATA4-regulated intestinal regionalization on bacterial colonization and tissue immunity, highlighting an elaborate interdependence of intestinal metabolism, immunity, and microbiota in homeostasis and disease.
Keywords: GATA4; IgA; bacterial colonization; celiac disease; immunopathology; infection; intestinal epithelial cells; intestinal regionalization; retinoic acid; segmented filamentous bacteria.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


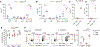

Comment in
-
GATA4 as a maestro of gut regionalization.Immunity. 2023 Jan 10;56(1):1-3. doi: 10.1016/j.immuni.2022.12.015. Immunity. 2023. PMID: 36630909
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

