An atlas of substrate specificities for the human serine/threonine kinome
- PMID: 36631611
- PMCID: PMC9876800
- DOI: 10.1038/s41586-022-05575-3
An atlas of substrate specificities for the human serine/threonine kinome
Abstract
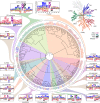
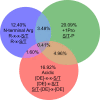
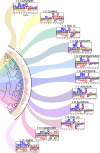
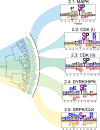
Protein phosphorylation is one of the most widespread post-translational modifications in biology1,2. With advances in mass-spectrometry-based phosphoproteomics, 90,000 sites of serine and threonine phosphorylation have so far been identified, and several thousand have been associated with human diseases and biological processes3,4. For the vast majority of phosphorylation events, it is not yet known which of the more than 300 protein serine/threonine (Ser/Thr) kinases encoded in the human genome are responsible3. Here we used synthetic peptide libraries to profile the substrate sequence specificity of 303 Ser/Thr kinases, comprising more than 84% of those predicted to be active in humans. Viewed in its entirety, the substrate specificity of the kinome was substantially more diverse than expected and was driven extensively by negative selectivity. We used our kinome-wide dataset to computationally annotate and identify the kinases capable of phosphorylating every reported phosphorylation site in the human Ser/Thr phosphoproteome. For the small minority of phosphosites for which the putative protein kinases involved have been previously reported, our predictions were in excellent agreement. When this approach was applied to examine the signalling response of tissues and cell lines to hormones, growth factors, targeted inhibitors and environmental or genetic perturbations, it revealed unexpected insights into pathway complexity and compensation. Overall, these studies reveal the intrinsic substrate specificity of the human Ser/Thr kinome, illuminate cellular signalling responses and provide a resource to link phosphorylation events to biological pathways.
© 2023. The Author(s).
Conflict of interest statement
L.C.C. is a founder and member of the board of directors of Agios Pharmaceuticals and is a founder and receives research support from Petra Pharmaceuticals; is listed as an inventor on a patent (WO2019232403A1, Weill Cornell Medicine) for combination therapy for PI3K-associated disease or disorder, and the identification of therapeutic interventions to improve response to PI3K inhibitors for cancer treatment; is a co-founder and shareholder in Faeth Therapeutics; has equity in and consults for Cell Signaling Technologies, Volastra, Larkspur and 1 Base Pharmaceuticals; and consults for Loxo-Lilly. M.B.Y receives research support from Cardiff Oncology. T.M.Y. is a co-founder and stockholder and is on the board of directors of DESTROKE, an early-stage start-up developing mobile technology for automated clinical stroke detection. J.L.J has received consulting fees from Scorpion Therapeutics and Volastra Therapeutics. O.E. is a founder and equity holder of Volastra Therapeutics and OneThree Biotech; is a member of the scientific advisory board of Owkin, Freenome, Genetic Intelligence, Acuamark and Champions Oncology; and receives research support from Eli Lilly, Janssen and Sanofi. D.J.T. is a member of the scientific advisory board at Dewpoint Therapeutics. A.D. is an equity holder of Denali Therapeutics; and receives research support from Interline Therapeutics. N.V. reports consulting activities for Novartis and is on the scientific advisory board of Heligenics. M.D.G. is a co-founder and shareholder of Faeth Therapeutics, which is developing dietary and pharmacological therapies for cancer; and has received speaking and/or consulting fees from Pfizer, Novartis, Scorpion Therapeutics and Faeth Therapeutics.
Figures









Comment in
-
Targets mapped for almost all human kinase enzymes.Nature. 2023 Jan;613(7945):637-638. doi: 10.1038/d41586-022-04583-7. Nature. 2023. PMID: 36631581 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

