Proteomic analysis of protein lysine 2-hydroxyisobutyrylation (Khib) in soybean leaves
- PMID: 36631736
- PMCID: PMC9835227
- DOI: 10.1186/s12870-022-04033-6
Proteomic analysis of protein lysine 2-hydroxyisobutyrylation (Khib) in soybean leaves
Abstract
Background: Protein lysine 2-hydroxyisobutyrylation (Khib) is a novel post-translational modification (PTM) discovered in cells or tissues of animals, microorganisms and plants in recent years. Proteome-wide identification of Khib-modified proteins has been performed in several plant species, suggesting that Khib-modified proteins are involved in a variety of biological processes and metabolic pathways. However, the protein Khib modification in soybean, a globally important legume crop that provides the rich source of plant protein and oil, remains unclear.
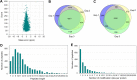
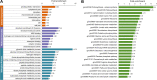
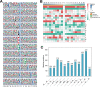
Results: In this study, the Khib-modified proteins in soybean leaves were identified for the first time using affinity enrichment and high-resolution mass spectrometry-based proteomic techniques, and a systematic bioinformatics analysis of these Khib-modified proteins was performed. Our results showed that a total of 4251 Khib sites in 1532 proteins were identified as overlapping in three replicates (the raw mass spectrometry data are available via ProteomeXchange with the identifier of PXD03650). These Khib-modified proteins are involved in a wide range of cellular processes, particularly enriched in biosynthesis, central carbon metabolism and photosynthesis, and are widely distributed in subcellular locations, mainly in chloroplasts, cytoplasm and nucleus. In addition, a total of 12 sequence motifs were extracted from all identified Khib peptides, and a basic amino acid residue (K), an acidic amino acid residue (E) and three aliphatic amino acid residues with small side chains (G/A/V) were found to be more preferred around the Khib site. Furthermore, 16 highly-connected clusters of Khib proteins were retrieved from the global PPI network, which suggest that Khib modifications tend to occur in proteins associated with specific functional clusters.
Conclusions: These findings suggest that Khib modification is an abundant and conserved PTM in soybean and that this modification may play an important role in regulating physiological processes in soybean leaves. The Khib proteomic data obtained in this study will help to further elucidate the regulatory mechanisms of Khib modification in soybean in the future.
Keywords: Lysine 2-hydroxyisobutyrylation (Khib); Motif; Post-translational modification (PTM); Proteomics; Soybean.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Global analysis of protein lysine 2-hydroxyisobutyrylation (Khib) profiles in Chinese herb rhubarb (Dahuang).BMC Genomics. 2021 Jul 15;22(1):542. doi: 10.1186/s12864-021-07847-0. BMC Genomics. 2021. PMID: 34266380 Free PMC article.
-
Systematic analysis of lysine 2-hydroxyisobutyrylation posttranslational modification in wheat leaves.PLoS One. 2021 Jun 17;16(6):e0253325. doi: 10.1371/journal.pone.0253325. eCollection 2021. PLoS One. 2021. PMID: 34138952 Free PMC article.
-
Proteome-wide analysis of lysine 2-hydroxyisobutyrylation in Frankliniella occidentalis.BMC Genomics. 2022 Aug 29;23(1):621. doi: 10.1186/s12864-022-08841-w. BMC Genomics. 2022. PMID: 36038823 Free PMC article.
-
Research advances in protein lysine 2-hydroxyisobutyrylation: From mechanistic regulation to disease relevance.J Cell Physiol. 2024 Dec;239(12):e31435. doi: 10.1002/jcp.31435. Epub 2024 Oct 1. J Cell Physiol. 2024. PMID: 39351825 Review.
-
Functions of lysine 2-hydroxyisobutyrylation and future perspectives on plants.Proteomics. 2023 Oct;23(19):e2300045. doi: 10.1002/pmic.202300045. Epub 2023 Jun 20. Proteomics. 2023. PMID: 37338329 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

