Genome-wide mapping of fluoroquinolone-stabilized DNA gyrase cleavage sites displays drug specific effects that correlate with bacterial persistence
- PMID: 36631985
- PMCID: PMC9943676
- DOI: 10.1093/nar/gkac1223
Genome-wide mapping of fluoroquinolone-stabilized DNA gyrase cleavage sites displays drug specific effects that correlate with bacterial persistence
Abstract
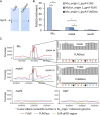
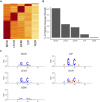
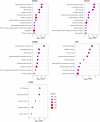
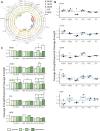
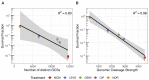
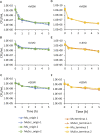
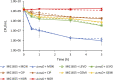
Bacterial persisters are rare phenotypic variants that are suspected to be culprits of recurrent infections. Fluoroquinolones (FQs) are a class of antibiotics that facilitate bacterial killing by stabilizing bacterial type II topoisomerases when they are in a complex with cleaved DNA. In Escherichia coli, DNA gyrase is the primary FQ target, and previous work has demonstrated that persisters are not spared from FQ-induced DNA damage. Since DNA gyrase cleavage sites (GCSs) largely govern the sites of DNA damage from FQ treatment, we hypothesized that GCS characteristics (e.g. number, strength, location) may influence persistence. To test this hypothesis, we measured genome-wide GCS distributions after treatment with a panel of FQs in stationary-phase cultures. We found drug-specific effects on the GCS distribution and discovered a strong negative correlation between the genomic cleavage strength and FQ persister levels. Further experiments and analyses suggested that persistence was unlikely to be governed by cleavage to individual sites, but rather survival was a function of the genomic GCS distribution. Together, these findings demonstrate FQ-specific differences in GCS distribution that correlate with persister levels and suggest that FQs that better stabilize DNA gyrase in cleaved complexes with DNA will lead to lower levels of persistence.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

