A computational approach to identify efficient RNA cleaving 10-23 DNAzymes
- PMID: 36632612
- PMCID: PMC9830538
- DOI: 10.1093/nargab/lqac098
A computational approach to identify efficient RNA cleaving 10-23 DNAzymes
Abstract
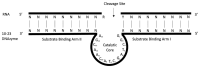
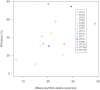
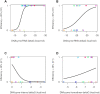
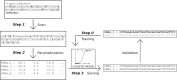
DNAzymes are short pieces of DNA with catalytic activity, capable of cleaving RNA. DNAzymes have multiple applications as biosensors and in therapeutics. The high specificity and low toxicity of these molecules make them particularly suitable as therapeutics, and clinical trials have shown that they are effective in patients. However, the development of DNAzymes has been limited due to the lack of specific tools to identify efficient molecules, and users often resort to time-consuming/costly large-scale screens. Here, we propose a computational methodology to identify 10-23 DNAzymes that can be used to triage thousands of potential molecules, specific to a target RNA, to identify those that are predicted to be efficient. The method is based on a logistic regression and can be trained to incorporate additional DNAzyme efficiency data, improving its performance with time. We first trained the method with published data, and then we validated, and further refined it, by testing additional newly synthesized DNAzymes in the laboratory. We found that although binding free energy between the DNAzyme and its RNA target is the primary determinant of efficiency, other factors such as internal structure of the DNAzyme also have an important effect. A program implementing the proposed method is publicly available.
© The Author(s) 2023. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures
Similar articles
-
Discovery and Biosensing Applications of Diverse RNA-Cleaving DNAzymes.Acc Chem Res. 2017 Sep 19;50(9):2273-2283. doi: 10.1021/acs.accounts.7b00262. Epub 2017 Aug 14. Acc Chem Res. 2017. PMID: 28805376
-
A complex RNA-cleaving DNAzyme that can efficiently cleave a pyrimidine-pyrimidine junction.J Mol Biol. 2010 Jul 23;400(4):689-701. doi: 10.1016/j.jmb.2010.05.047. Epub 2010 May 26. J Mol Biol. 2010. PMID: 20630470
-
In vitro selection of RNA-cleaving DNAzymes for bacterial detection.Methods. 2016 Aug 15;106:66-75. doi: 10.1016/j.ymeth.2016.03.018. Epub 2016 Mar 24. Methods. 2016. PMID: 27017912
-
Theranostic DNAzymes.Theranostics. 2017 Feb 23;7(4):1010-1025. doi: 10.7150/thno.17736. eCollection 2017. Theranostics. 2017. PMID: 28382172 Free PMC article. Review.
-
DNAzyme-based biosensors for mercury (Ⅱ) detection: Rational construction, advances and perspectives.J Hazard Mater. 2022 Jun 5;431:128606. doi: 10.1016/j.jhazmat.2022.128606. Epub 2022 Mar 3. J Hazard Mater. 2022. PMID: 35278952 Review.
Cited by
-
SequenceCraft: machine learning-based resource for exploratory analysis of RNA-cleaving deoxyribozymes.BMC Bioinformatics. 2025 Jan 6;26(1):2. doi: 10.1186/s12859-024-06019-7. BMC Bioinformatics. 2025. PMID: 39762738 Free PMC article.
-
Novel In Vitro Selection of Trans-Acting BCL-2 mRNA-Cleaving Deoxyribozymes for Cancer Therapy.Cells. 2025 Jun 20;14(13):945. doi: 10.3390/cells14130945. Cells. 2025. PMID: 40643466 Free PMC article.
-
Making target sites in large structured RNAs accessible to RNA-cleaving DNAzymes through hybridization with synthetic DNA oligonucleotides.Nucleic Acids Res. 2024 Oct 14;52(18):11177-11187. doi: 10.1093/nar/gkae778. Nucleic Acids Res. 2024. PMID: 39248110 Free PMC article.
References
-
- Breaker R.R., Joyce G.F.. A DNA enzyme that cleaves RNA. Chem. Biol. 1994; 1:223–229. - PubMed
-
- Morrison D., Rothenbroker M., Li Y.. DNAzymes: selected for applications. Small Methods. 2018; 2:1700319.
-
- Ali M.M., Wolfe M., Tram K., Gu J., Filipe C.D.M., Li Y., Brennan J.D.. A DNAzyme-based colorimetric paper sensor for Helicobacter pylori. Angew. Chem. Int. Ed. 2019; 58:9907–9911. - PubMed
LinkOut - more resources
Full Text Sources

