Targeting the secreted RGDKGE collagen fragment reduces PD‑L1 by a proteasome‑dependent mechanism and inhibits tumor growth
- PMID: 36633146
- PMCID: PMC9868893
- DOI: 10.3892/or.2023.8481
Targeting the secreted RGDKGE collagen fragment reduces PD‑L1 by a proteasome‑dependent mechanism and inhibits tumor growth
Abstract
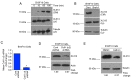
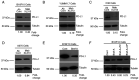
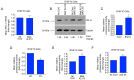
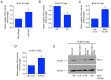
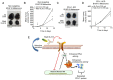
Structural alterations of collagen impact signaling that helps control tumor progression and the responses to therapeutic intervention. Integrins represent a class of receptors that include members that mediate collagen signaling. However, a strategy of directly targeting integrins to control tumor growth has demonstrated limited activity in the clinical setting. New molecular understanding of integrins have revealed that these receptors can regulate both pro‑ and anti‑tumorigenic functions in a cell type‑dependent manner. Therefore, designing strategies that block pro‑tumorigenic signaling, without impeding anti‑tumorigenic functions, may lead to development of more effective therapies. In the present study, evidence was provided for a novel signaling cascade in which β3‑integrin‑mediated binding to a secreted RGDKGE‑containing collagen fragment stimulates an autocrine‑like signaling pathway that differentially governs the activity of both YAP and (protein kinase‑A) PKA, ultimately leading to alterations in the levels of immune checkpoint molecule PD‑L1 by a proteasome dependent mechanism. Selectively targeting this collagen fragment, reduced nuclear YAP levels, and enhanced PKA and proteasome activity, while also exhibiting significant antitumor activity in vivo. The present findings not only provided new mechanistic insight into a previously unknown autocrine‑like signaling pathway that may provide tumor cells with the ability to regulate PD‑L1, but our findings may also help in the development of more effective strategies to control pro‑tumorigenic β3‑integrin signaling without disrupting its tumor suppressive functions in other cellular compartments.
Keywords: YAP; collagen; extracellular matrix; integrin αvβ3; programmed death‑ligand 1; protein kinase‑A; stroma.
Conflict of interest statement
PCB holds an equity position in CryptoMedix, Inc. The rest of the authors declare that they have no competing interests.
Figures


Similar articles
-
An RGDKGE-Containing Cryptic Collagen Fragment Regulates Phosphorylation of Large Tumor Suppressor Kinase-1 and Controls Ovarian Tumor Growth by a Yes-Associated Protein-Dependent Mechanism.Am J Pathol. 2021 Mar;191(3):527-544. doi: 10.1016/j.ajpath.2020.11.009. Epub 2020 Dec 8. Am J Pathol. 2021. PMID: 33307038 Free PMC article.
-
Identification of an Endogenously Generated Cryptic Collagen Epitope (XL313) That May Selectively Regulate Angiogenesis by an Integrin Yes-associated Protein (YAP) Mechano-transduction Pathway.J Biol Chem. 2016 Feb 5;291(6):2731-50. doi: 10.1074/jbc.M115.669614. Epub 2015 Dec 14. J Biol Chem. 2016. PMID: 26668310 Free PMC article.
-
Targeting of interleukin (IL)-17A inhibits PDL1 expression in tumor cells and induces anticancer immunity in an estrogen receptor-negative murine model of breast cancer.Oncotarget. 2017 Jan 31;8(5):7614-7624. doi: 10.18632/oncotarget.13819. Oncotarget. 2017. PMID: 27935862 Free PMC article.
-
Collagen remodeling-mediated signaling pathways and their impact on tumor therapy.J Biol Chem. 2025 Mar;301(3):108330. doi: 10.1016/j.jbc.2025.108330. Epub 2025 Feb 19. J Biol Chem. 2025. PMID: 39984051 Free PMC article. Review.
-
What role does PDL1 play in EMT changes in tumors and fibrosis?Front Immunol. 2023 Aug 15;14:1226038. doi: 10.3389/fimmu.2023.1226038. eCollection 2023. Front Immunol. 2023. PMID: 37649487 Free PMC article. Review.
Cited by
-
Glycosylation in the tumor immune response: the bitter side of sweetness.Acta Biochim Biophys Sin (Shanghai). 2024 Jun 28;56(8):1184-1198. doi: 10.3724/abbs.2024107. Acta Biochim Biophys Sin (Shanghai). 2024. PMID: 38946426 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

