Discovery, implications and initial use of semi-synthetic organisms with an expanded genetic alphabet/code
- PMID: 36633274
- PMCID: PMC9835597
- DOI: 10.1098/rstb.2022.0030
Discovery, implications and initial use of semi-synthetic organisms with an expanded genetic alphabet/code
Abstract
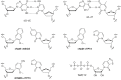
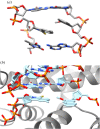
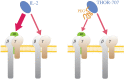
Much recent interest has focused on developing proteins for human use, such as in medicine. However, natural proteins are made up of only a limited number of canonical amino acids with limited functionalities, and this makes the discovery of variants with some functions difficult. The ability to recombinantly express proteins containing non-canonical amino acids (ncAAs) with properties selected to impart the protein with desired properties is expected to dramatically improve the discovery of proteins with different functions. Perhaps the most straightforward approach to such an expansion of the genetic code is through expansion of the genetic alphabet, so that new codon/anticodon pairs can be created to assign to ncAAs. In this review, I briefly summarize more than 20 years of effort leading ultimately to the discovery of synthetic nucleotides that pair to form an unnatural base pair, which when incorporated into DNA, is stably maintained, transcribed and used to translate proteins in Escherichia coli. In addition to discussing wide ranging conceptual implications, I also describe ongoing efforts at the pharmaceutical company Sanofi to employ the resulting 'semi-synthetic organisms' or SSOs, for the production of next-generation protein therapeutics. This article is part of the theme issue 'Reactivity and mechanism in chemical and synthetic biology'.
Keywords: expanded genetic alphabet; expanded genetic code; protein therapeutics; semi-synthetic organisms; unnatural base pair.
Figures


Similar articles
-
Creation, Optimization, and Use of Semi-Synthetic Organisms that Store and Retrieve Increased Genetic Information.J Mol Biol. 2022 Apr 30;434(8):167331. doi: 10.1016/j.jmb.2021.167331. Epub 2021 Oct 25. J Mol Biol. 2022. PMID: 34710400 Review.
-
Expansion of the genetic code via expansion of the genetic alphabet.Curr Opin Chem Biol. 2018 Oct;46:196-202. doi: 10.1016/j.cbpa.2018.08.009. Epub 2018 Sep 8. Curr Opin Chem Biol. 2018. PMID: 30205312 Free PMC article. Review.
-
Expansion of the Genetic Alphabet: A Chemist's Approach to Synthetic Biology.Acc Chem Res. 2018 Feb 20;51(2):394-403. doi: 10.1021/acs.accounts.7b00403. Epub 2017 Dec 2. Acc Chem Res. 2018. PMID: 29198111 Free PMC article.
-
New codons for efficient production of unnatural proteins in a semisynthetic organism.Nat Chem Biol. 2020 May;16(5):570-576. doi: 10.1038/s41589-020-0507-z. Epub 2020 Apr 6. Nat Chem Biol. 2020. PMID: 32251411 Free PMC article.
-
A semi-synthetic organism with an expanded genetic alphabet.Nature. 2014 May 15;509(7500):385-8. doi: 10.1038/nature13314. Epub 2014 May 7. Nature. 2014. PMID: 24805238 Free PMC article.
Cited by
-
Cellular Site-Specific Incorporation of Noncanonical Amino Acids in Synthetic Biology.Chem Rev. 2024 Sep 25;124(18):10577-10617. doi: 10.1021/acs.chemrev.3c00938. Epub 2024 Aug 29. Chem Rev. 2024. PMID: 39207844 Review.
-
Reactivity and mechanism in chemical and synthetic biology.Philos Trans R Soc Lond B Biol Sci. 2023 Feb 27;378(1871):20220023. doi: 10.1098/rstb.2022.0023. Epub 2023 Jan 11. Philos Trans R Soc Lond B Biol Sci. 2023. PMID: 36633278 Free PMC article.
-
A Prebiotic Genetic Nucleotide as an Early Darwinian Ancestor for Pre-RNA Evolution.ACS Omega. 2024 Apr 12;9(16):18072-18082. doi: 10.1021/acsomega.3c09949. eCollection 2024 Apr 23. ACS Omega. 2024. PMID: 38680342 Free PMC article.
-
Residue-Specific Incorporation of Noncanonical Amino Acids in Auxotrophic Hosts: Quo Vadis?.Chem Rev. 2025 May 28;125(10):4840-4932. doi: 10.1021/acs.chemrev.4c00280. Epub 2025 May 16. Chem Rev. 2025. PMID: 40378355 Free PMC article. Review.
-
Genetic Code Expansion: Recent Developments and Emerging Applications.Chem Rev. 2025 Jan 22;125(2):523-598. doi: 10.1021/acs.chemrev.4c00216. Epub 2024 Dec 31. Chem Rev. 2025. PMID: 39737807 Free PMC article. Review.
References
-
- Wang L, Magliery TJ, Liu DR, Schultz PG. 2000. A new functional suppressor tRNA/aminoacyl–tRNA synthetase pair for the in vivo incorporation of unnatural amino acids into proteins. J. Am. Chem. Soc. 122, 5010-5011. (10.1021/ja000595y) - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

