Structure, mechanism and inhibition of anthranilate phosphoribosyltransferase
- PMID: 36633281
- PMCID: PMC9835598
- DOI: 10.1098/rstb.2022.0039
Structure, mechanism and inhibition of anthranilate phosphoribosyltransferase
Abstract
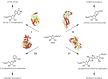
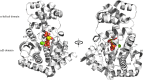
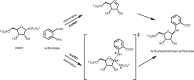
Anthranilate phosphoribosyltransferase catalyses the second reaction in the biosynthesis of tryptophan from chorismate in microorganisms and plants. The enzyme is homodimeric with the active site located in the hinge region between two domains. A range of structures in complex with the substrates, substrate analogues and inhibitors have been determined, and these have provided insights into the catalytic mechanism of this enzyme. Substrate 5-phospho-d-ribose 1-diphosphate (PRPP) binds to the C-terminal domain and coordinates to Mg2+, in a site completed by two flexible loops. Binding of the second substrate anthranilate is more complex, featuring multiple binding sites along an anthranilate channel. This multi-modal binding is consistent with the substrate inhibition observed at high concentrations of anthranilate. A series of structures predict a dissociative mechanism for the reaction, similar to the reaction mechanisms elucidated for other phosphoribosyltransferases. As this enzyme is essential for some pathogens, efforts have been made to develop inhibitors for this enzyme. To date, the best inhibitors exploit the multiple binding sites for anthranilate. This article is part of the theme issue 'Reactivity and mechanism in chemical and synthetic biology'.
Keywords: AnPRT; PRT; TrpD; tryptophan biosynthesis.
Conflict of interest statement
We declare we have no competing interests.
Figures
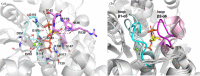


Similar articles
-
The substrate capture mechanism of Mycobacterium tuberculosis anthranilate phosphoribosyltransferase provides a mode for inhibition.Biochemistry. 2013 Mar 12;52(10):1776-87. doi: 10.1021/bi301387m. Epub 2013 Feb 28. Biochemistry. 2013. PMID: 23363292
-
Alternative substrates reveal catalytic cycle and key binding events in the reaction catalysed by anthranilate phosphoribosyltransferase from Mycobacterium tuberculosis.Biochem J. 2014 Jul 1;461(1):87-98. doi: 10.1042/BJ20140209. Biochem J. 2014. PMID: 24712732
-
The crystal structure of TrpD, a metabolic enzyme essential for lung colonization by Mycobacterium tuberculosis, in complex with its substrate phosphoribosylpyrophosphate.J Mol Biol. 2006 Jan 27;355(4):784-97. doi: 10.1016/j.jmb.2005.11.016. Epub 2005 Nov 22. J Mol Biol. 2006. PMID: 16337227
-
Anthranilate phosphoribosyltransferase: Binding determinants for 5'-phospho-alpha-d-ribosyl-1'-pyrophosphate (PRPP) and the implications for inhibitor design.Biochim Biophys Acta Proteins Proteom. 2018 Feb;1866(2):264-274. doi: 10.1016/j.bbapap.2017.08.018. Epub 2017 Aug 26. Biochim Biophys Acta Proteins Proteom. 2018. PMID: 28844746
-
Structural features of the phosphoribosyltransferases and their relationship to the human deficiency disorders of purine and pyrimidine metabolism.CRC Crit Rev Biochem. 1981;11(1):1-34. doi: 10.3109/10409238109108698. CRC Crit Rev Biochem. 1981. PMID: 7030616 Review.
Cited by
-
Phosphoribosyltransferases and Their Roles in Plant Development and Abiotic Stress Response.Int J Mol Sci. 2023 Jul 23;24(14):11828. doi: 10.3390/ijms241411828. Int J Mol Sci. 2023. PMID: 37511586 Free PMC article. Review.
-
Microbial colonization programs are structured by breastfeeding and guide healthy respiratory development.Cell. 2024 Sep 19;187(19):5431-5452.e20. doi: 10.1016/j.cell.2024.07.022. Cell. 2024. PMID: 39303691
-
Structure-Based Virtual Screening of Potential Inhibitors Targeting the Prolyl-tRNA Synthetase (PRS) in Eimeria tenella: Insights from Molecular Docking, ADMET Studies, and Molecular Dynamics Simulations.Molecules. 2025 Feb 8;30(4):790. doi: 10.3390/molecules30040790. Molecules. 2025. PMID: 40005102 Free PMC article.
-
Fermentative aminopyrrolnitrin production by metabolically engineered Corynebacterium glutamicum.Microb Cell Fact. 2024 May 23;23(1):147. doi: 10.1186/s12934-024-02424-y. Microb Cell Fact. 2024. PMID: 38783320 Free PMC article.
-
Reactivity and mechanism in chemical and synthetic biology.Philos Trans R Soc Lond B Biol Sci. 2023 Feb 27;378(1871):20220023. doi: 10.1098/rstb.2022.0023. Epub 2023 Jan 11. Philos Trans R Soc Lond B Biol Sci. 2023. PMID: 36633278 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

