Isolation and Characterization of Distinct Rotavirus A in Bat and Rodent Hosts
- PMID: 36633410
- PMCID: PMC9888233
- DOI: 10.1128/jvi.01455-22
Isolation and Characterization of Distinct Rotavirus A in Bat and Rodent Hosts
Abstract
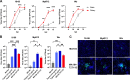
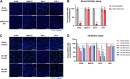
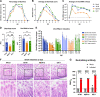
Rotavirus A (RVA) causes diarrheal disease in humans and various animals. Recent studies have identified bat and rodent RVAs with evidence of zoonotic transmission and genome reassortment. However, the virological properties of bat and rodent RVAs with currently identified genotypes still need to be better clarified. Here, we performed virus isolation-based screening for RVA in animal specimens and isolated RVAs (representative strains: 16-06 and MpR12) from Egyptian fruit bat and Natal multimammate mouse collected in Zambia. Whole-genome sequencing and phylogenetic analysis revealed that the genotypes of bat RVA 16-06 were identical to that of RVA BATp39 strain from the Kenyan fruit bat, which has not yet been characterized. Moreover, all segments of rodent RVA MpR12 were highly divergent and assigned to novel genotypes, but RVA MpR12 was phylogenetically closer to bat RVAs than to other rodent RVAs, indicating a unique evolutionary history. We further investigated the virological properties of the isolated RVAs. In brief, we found that 16-06 entered cells by binding to sialic acids on the cell surface, while MpR12 entered in a sialic acid-independent manner. Experimental inoculation of suckling mice with 16-06 and MpR12 revealed that these RVAs are causative agents of diarrhea. Moreover, 16-06 and MpR12 demonstrated an ability to infect and replicate in a 3D-reconstructed primary human intestinal epithelium with comparable efficiency to the human RVA. Taken together, our results detail the unique genetic and virological features of bat and rodent RVAs and demonstrate the need for further investigation of their zoonotic potential. IMPORTANCE Recent advances in nucleotide sequence detection methods have enabled the detection of RVA genomes from various animals. These studies have discovered multiple divergent RVAs and have resulted in proposals for the genetic classification of novel genotypes. However, most of these RVAs have been identified via dsRNA viral genomes and not from infectious viruses, and their virological properties, such as cell/host tropisms, transmissibility, and pathogenicity, are unclear and remain to be clarified. Here, we successfully isolated RVAs with novel genome constellations from three bats and one rodent in Zambia. In addition to whole-genome sequencing, the isolated RVAs were characterized by glycan-binding affinity, pathogenicity in mice, and infectivity to the human gut using a 3D culture of primary intestinal epithelium. Our study reveals the first virological properties of bat and rodent RVAs with high genetic diversity and unique evolutional history and provides basic knowledge to begin estimating the potential of zoonotic transmission.
Keywords: Rotavirus A; bats; genetic diversity; glycan specificity; human intestinal epithelium model; rodents.
Conflict of interest statement
The authors declare a conflict of interest. Shinsuke Toba is an employee of Shionogi & Co., Ltd. Other authors declare no competing interests.
Figures



Similar articles
-
Group A Rotaviruses in Chinese Bats: Genetic Composition, Serology, and Evidence for Bat-to-Human Transmission and Reassortment.J Virol. 2017 May 26;91(12):e02493-16. doi: 10.1128/JVI.02493-16. Print 2017 Jun 15. J Virol. 2017. PMID: 28381569 Free PMC article.
-
At Least Seven Distinct Rotavirus Genotype Constellations in Bats with Evidence of Reassortment and Zoonotic Transmissions.mBio. 2021 Jan 19;12(1):e02755-20. doi: 10.1128/mBio.02755-20. mBio. 2021. PMID: 33468689 Free PMC article.
-
Complete Genome Sequencing Reveals Unusual Equine Rotavirus A of Bat Origin from India.J Virol. 2022 Oct 26;96(20):e0140822. doi: 10.1128/jvi.01408-22. Epub 2022 Oct 10. J Virol. 2022. PMID: 36214578 Free PMC article.
-
Unusual G3P[10] bat-like rotavirus strains detected in children with acute gastroenteritis in Thailand.J Med Virol. 2024 Oct;96(10):e70014. doi: 10.1002/jmv.70014. J Med Virol. 2024. PMID: 39420695
-
Whole genome analysis provides evidence for porcine-to-simian interspecies transmission of rotavirus-A.Infect Genet Evol. 2017 Apr;49:21-31. doi: 10.1016/j.meegid.2016.12.026. Epub 2016 Dec 28. Infect Genet Evol. 2017. PMID: 28039076
Cited by
-
Emergence of a Novel G4P[6] Porcine Rotavirus with Unique Sequence Duplication in NSP5 Gene in China.Animals (Basel). 2024 Jun 14;14(12):1790. doi: 10.3390/ani14121790. Animals (Basel). 2024. PMID: 38929409 Free PMC article.
-
Diversity and Potential Cross-Species Transmission of Rotavirus A in Wild Animals in Yunnan, China.Microorganisms. 2025 Jan 13;13(1):145. doi: 10.3390/microorganisms13010145. Microorganisms. 2025. PMID: 39858913 Free PMC article.
-
Genetic and antigenic characterization of two diarrhoeicdominant rotavirus A genotypes G3P[12] and G14P[12] circulating in the global equine population.J Gen Virol. 2024 Aug;105(8):002016. doi: 10.1099/jgv.0.002016. J Gen Virol. 2024. PMID: 39163114 Free PMC article.
-
Prevalence and Genomic Characterization of Rotavirus A from Domestic Pigs in Zambia: Evidence for Possible Porcine-Human Interspecies Transmission.Pathogens. 2023 Sep 27;12(10):1199. doi: 10.3390/pathogens12101199. Pathogens. 2023. PMID: 37887715 Free PMC article.
-
Isolation and Genomic Characterization of the G6P[1]-Type Sheep Rotavirus in China.Transbound Emerg Dis. 2024 Jul 1;2024:9614599. doi: 10.1155/2024/9614599. eCollection 2024. Transbound Emerg Dis. 2024. PMID: 40303190 Free PMC article.
References
-
- Sadiq A, Bostan N, Khan J, Aziz A. 2022. Effect of rotavirus genetic diversity on vaccine impact. Rev Med Virol 32:e2259. - PubMed
-
- Malik YS, Bhat S, Dar PS, Sircar S, Dhama K, Singh RK. 2020. Evolving rotaviruses, interspecies transmission and zoonoses. Open Virol J 14:1–6. 10.2174/1874357902014010001. - DOI
-
- Matthijnssens J, Ciarlet M, Heiman E, Arijs I, Delbeke T, McDonald SM, Palombo EA, Iturriza-Gómara M, Maes P, Patton JT, Rahman M, Van Ranst M. 2008. Full genome-based classification of rotaviruses reveals a common origin between human Wa-like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J Virol 82:3204–3219. 10.1128/JVI.02257-07. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

