15 years of GWAS discovery: Realizing the promise
- PMID: 36634672
- PMCID: PMC9943775
- DOI: 10.1016/j.ajhg.2022.12.011
15 years of GWAS discovery: Realizing the promise
Abstract
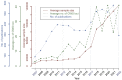
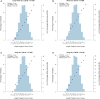
It has been 15 years since the advent of the genome-wide association study (GWAS) era. Here, we review how this experimental design has realized its promise by facilitating an impressive range of discoveries with remarkable impact on multiple fields, including population genetics, complex trait genetics, epidemiology, social science, and medicine. We predict that the emergence of large-scale biobanks will continue to expand to more diverse populations and capture more of the allele frequency spectrum through whole-genome sequencing, which will further improve our ability to investigate the causes and consequences of human genetic variation for complex traits and diseases.
Copyright © 2022 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

References
-
- Risch N., Merikangas K. The future of genetic studies of complex human diseases. Science (New York, N.Y.) 1996;273:1516–1517. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

