Regulators of male and female sexual development are critical for the transmission of a malaria parasite
- PMID: 36634679
- PMCID: PMC7616090
- DOI: 10.1016/j.chom.2022.12.011
Regulators of male and female sexual development are critical for the transmission of a malaria parasite
Abstract
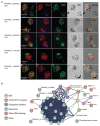
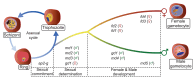
Malaria transmission to mosquitoes requires a developmental switch in asexually dividing blood-stage parasites to sexual reproduction. In Plasmodium berghei, the transcription factor AP2-G is required and sufficient for this switch, but how a particular sex is determined in a haploid parasite remains unknown. Using a global screen of barcoded mutants, we here identify genes essential for the formation of either male or female sexual forms and validate their importance for transmission. High-resolution single-cell transcriptomics of ten mutant parasites portrays the developmental bifurcation and reveals a regulatory cascade of putative gene functions in the determination and subsequent differentiation of each sex. A male-determining gene with a LOTUS/OST-HTH domain as well as the protein interactors of a female-determining zinc-finger protein indicate that germ-granule-like ribonucleoprotein complexes complement transcriptional processes in the regulation of both male and female development of a malaria parasite.
Keywords: Plasmodium; development; differentiation; malaria; sex determination; sex ratio; single cell analysis; transmission.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






Comment in
-
The troubled puberty of malaria parasites.Trends Parasitol. 2023 Mar;39(3):155-157. doi: 10.1016/j.pt.2023.01.006. Epub 2023 Jan 24. Trends Parasitol. 2023. PMID: 36702699
-
Towards a general, worldwide, Plasmodium population genomics framework.Trends Parasitol. 2023 Apr;39(4):229-230. doi: 10.1016/j.pt.2023.01.003. Epub 2023 Jan 25. Trends Parasitol. 2023. PMID: 36707341 No abstract available.
References
-
- Brancucci NMB, Bertschi NL, Zhu L, Niederwieser I, Chin WH, Wampfler R, Freymond C, Rottmann M, Felger I, Bozdech Z, Voss TS. Heterochromatin protein 1 secures survival and transmission of malaria parasites. Cell Host Microbe. 2014;16:165–176. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

