Targeting TBK1 to overcome resistance to cancer immunotherapy
- PMID: 36634707
- PMCID: PMC10171827
- DOI: 10.1038/s41586-023-05704-6
Targeting TBK1 to overcome resistance to cancer immunotherapy
Abstract
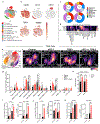
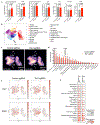
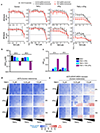
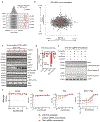
Despite the success of PD-1 blockade in melanoma and other cancers, effective treatment strategies to overcome resistance to cancer immunotherapy are lacking1,2. Here we identify the innate immune kinase TANK-binding kinase 1 (TBK1)3 as a candidate immune-evasion gene in a pooled genetic screen4. Using a suite of genetic and pharmacological tools across multiple experimental model systems, we confirm a role for TBK1 as an immune-evasion gene. Targeting TBK1 enhances responses to PD-1 blockade by decreasing the cytotoxicity threshold to effector cytokines (TNF and IFNγ). TBK1 inhibition in combination with PD-1 blockade also demonstrated efficacy using patient-derived tumour models, with concordant findings in matched patient-derived organotypic tumour spheroids and matched patient-derived organoids. Tumour cells lacking TBK1 are primed to undergo RIPK- and caspase-dependent cell death in response to TNF and IFNγ in a JAK-STAT-dependent manner. Taken together, our results demonstrate that targeting TBK1 is an effective strategy to overcome resistance to cancer immunotherapy.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
R.W.J. is a member of the advisory board for and has a financial interest in Xsphera Biosciences Inc., a company focused on using ex vivo profiling technology to deliver functional, precision immune-oncology solutions for patients, providers, and drug development companies. A.M. is a consultant for Third Rock Ventures, LLC., Asher Biotherapeutics and Abata Therapeutics. A.M. holds equity in Asher Biotherapeutics and Abata Therapeutics. S.I.P. has received consultancy payments from Abbvie, Astrazeneca/MedImmune, Cue Biopharma, Fusion Pharmaceuticals, MSD/Merck, Newlink Genetics, Oncolys Biopharma, Replimmune, Scopus Biopharma, and Sensei Biopharma. She has received grants/research support from Abbvie, Astrazeneca/MedImmune, Cue Biopharma, Merck, and Tesaro. S.J.K. has served a consultant/advisory role for Eli Lilly, Merck, BMS, Astellas, Daiichi-Sankyo, Pieris, and Natera. S.J.K. owns stock in Turning Point Therapeutics, Inc. D.J. received consulting fees from Novartis, Genentech, Syros, Eisai, Vibliome, Mapkure, and Relay Therapeutics. D.J. conducted contracted research with Novartis, Genentech, Syros, Pfizer, Eisai, Takeda, Pfizer, Ribon Therapeutics, Infinity, InventisBio, and Arvinas. D.J. has ownership interest in Relay Therapeutics and PIC Therapeutics. D.M.M. has received honoraria for participating on advisory boards for Checkpoint Therapeutics, EMD Serono, Castle Biosciences, Pfizer, Merck, Regeneron, and Sanofi Genzyme. D.M.M. owns stock for Checkpoint Therapeutics. D.E.F. has a financial interest in Soltego, a company developing salt inducible kinase inhibitors for topical skin-darkening treatments that might be used for a broad set of human applications. R.T.M. consults for Bristol Myers Squibb. M.S.F receives research funding from Bristol-Meyers Squib.
Figures











References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

